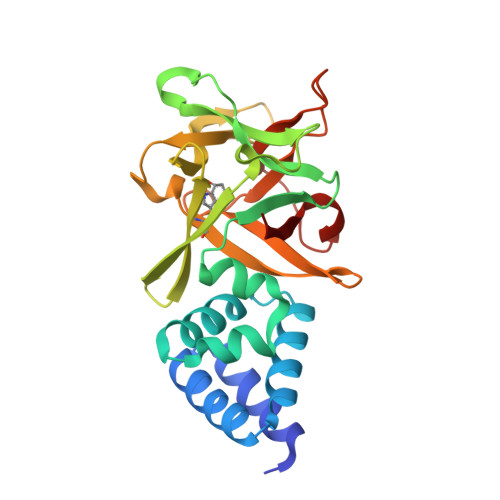
High-resolution structure of Bombyx mori lipoprotein 7: crystallographic determination of the identity of the protein and its potential role in detoxification.
Pietrzyk, A.J., Panjikar, S., Bujacz, A., Mueller-Dieckmann, J., Lochynska, M., Jaskolski, M., Bujacz, G.(2012) Acta Crystallogr D Biol Crystallogr 68: 1140-1151
- PubMed: 22948915
- DOI: https://doi.org/10.1107/S0907444912021555
- Primary Citation of Related Structures:
4EFP, 4EFQ, 4EFR - PubMed Abstract:
Three crystal structures of a lipoprotein (Bmlp7) of unknown function, a member of the 30 kDa lipoprotein family from mulberry silkworm (Bombyx mori L.) haemolymph, have been determined. The 1.33 Å resolution structure is an excellent example of how a precise crystallographic study can contribute to protein identification. The correct sequence of this haemolymph-isolated protein was assigned thanks to superb-quality electron-density maps. Two unexpected cadmium cations were found in this crystal structure [Bmlp7-I(Cd)] and their presence may be connected to a detoxification mechanism in this insect. For a comparison of the metal-binding sites, the crystal structure of a platinum complex (Bmlp7-Pt) was also solved at 1.94 Å resolution. The third (2.50 Å resolution) structure, of the native protein harvested in a different season (Bmlp7-II), corresponds to a different polymorph with an altered pattern of intermolecular interactions and with a total absence of cadmium ions and highlights the possible involvement of Bmlp7 in the response to environmental pollution. The N-terminal domain of Bmlp7 has a fold resembling a clockwise spiral created by six helices and can be classified as a VHS domain. The C-terminal domain is folded as a β-trefoil. The biological function of Bmlp7 is unknown, but its structural homology to sugar-binding proteins suggests that, in analogy to other 30 kDa haemolymph lipoproteins, it could play a role as an anti-apoptotic factor or function in the immune response of the insect to fungal infections.
Organizational Affiliation:
Center for Biocrystallographic Research, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland.