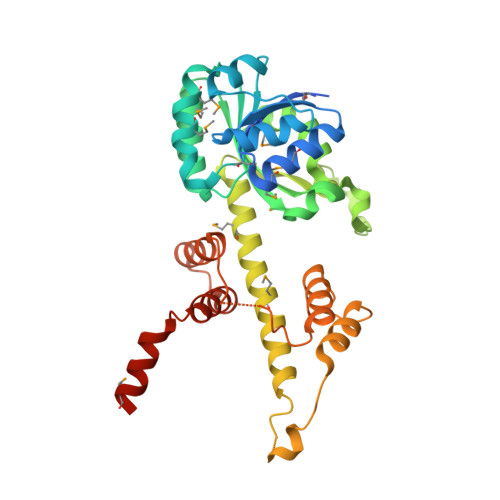
The crystal structure of 6-phosphogluconate dehydrogenase from Geobacter metallireducens
Zhang, Z., Chamala, S., Evans, B., Foti, R., Gizzi, A., Hillerich, B., Kar, A., Lafleur, J., Seidel, R., Villigas, G., Zencheck, W., Almo, S.C., Swaminathan, S.To be published.