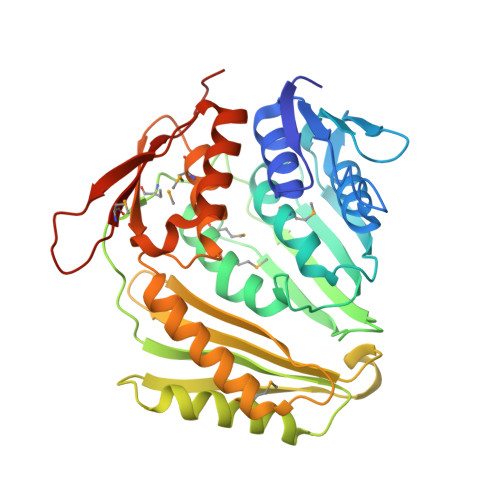
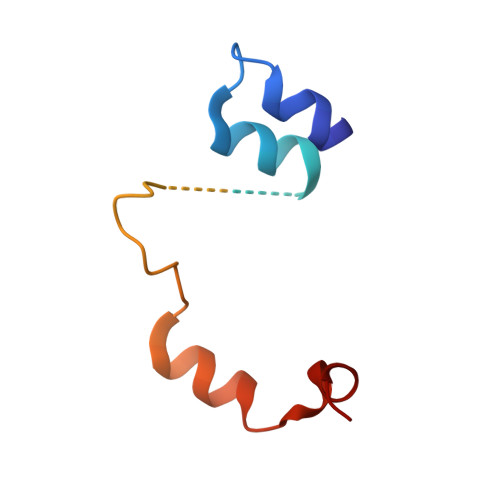
Crucial Role of the Rcl1P-Bms1P Interaction for Yeast Pre-Ribosomal RNA Processing.
Delprato, A., Al Kadri, Y., Perebaskine, N., Monfoulet, C., Henry, Y., Henras, A.K., Fribourg, S.(2014) Nucleic Acids Res 42: 10161
- PubMed: 25064857
- DOI: https://doi.org/10.1093/nar/gku682
- Primary Citation of Related Structures:
4CLQ - PubMed Abstract:
The essential Rcl1p and Bms1p proteins form a complex required for 40S ribosomal subunit maturation. Bms1p is a GTPase and Rcl1p has been proposed to catalyse the endonucleolytic cleavage at site A2 separating the pre-40S and pre-60S maturation pathways. We determined the 2.0 Å crystal structure of Bms1p associated with Rcl1p. We demonstrate that Rcl1p nuclear import depends on Bms1p and that the two proteins are loaded into pre-ribosomes at a similar stage of the maturation pathway and remain present within pre-ribosomes after cleavage at A2. Importantly, GTP binding to Bms1p is not required for the import in the nucleus nor for the incorporation of Rcl1p into pre-ribosomes, but is essential for early pre-rRNA processing. We propose that GTP binding to Bms1p and/or GTP hydrolysis may induce conformational rearrangements within the Bms1p-Rcl1p complex allowing the interaction of Rcl1p with its RNA substrate.
Organizational Affiliation:
Institut Européen de Chimie et Biologie, ARNA laboratory, Université de Bordeaux, F-33607 Pessac, France Institut National de la Santé Et de la Recherche Médicale, INSERM - U869, ARNA laboratory, F-33000 Bordeaux, France.