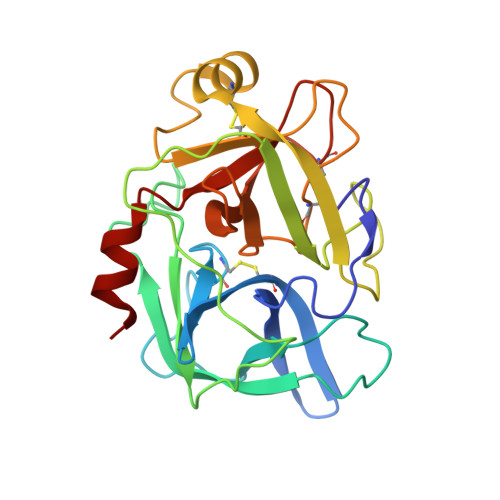
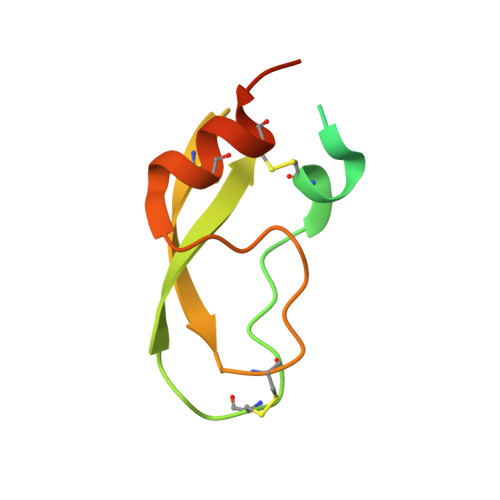
Comparison of Complexes Formed by a Crustacean and a Vertebrate Trypsin with Bovine Pancreatic Trypsin Inhibitor - the Key to Achieving Extreme Stability?
Molnar, T., Voros, J., Szeder, B., Takats, K., Kardos, J., Katona, G., Graf, L.(2013) FEBS J 280: 5750
- PubMed: 24034223
- DOI: https://doi.org/10.1111/febs.12491
- Primary Citation of Related Structures:
4BNR - PubMed Abstract:
This paper provides evidence for the extremely high resistance of a complex of crayfish trypsin (CFT) and bovine pancreatic trypsin inhibitor (BPTI) against heating and chemical denaturing agents such as sodium dodecyl sulfate (SDS) and urea. To dissociate this complex, 15 min boiling in SDS was necessary, compared to a complex of bovine trypsin (BT) (EC 3.4.21.4) and BPTI, which dissociates in SDS without boiling. The CFT-BPTI complex remained stable even in 9 m urea, while the BT-BPTI complex started to dissociate at concentrations of approximately 4 m urea. The melting temperatures of the BT-BPTI and CFT-BPTI complexes, as determined by differential scanning calorimetry, were found to be 79.6 and 100.1 °C, respectively. The behaviour of the apo-enzymes - CFT was found to have a less stable structure compared to BT - did not provide a definite indication regarding the differential effects on their stabilities. To explore the structural features responsible for this extreme stability, we crystallized CFT in complex with BPTI, and identified extended contacts compared to the BT-BPTI complex. Comparison of the B-factors of similar trypsin-trypsin inhibitor complexes suggests that molecular flexibility of the components is also required for the strong protein-protein interaction. Although the structural reason for the extreme stability of the CFT-BPTI complex is not yet fully understood, our study may be a starting point for the development of new protein complexes with enhanced stability.
Organizational Affiliation:
Department of Biochemistry, Eötvös Loránd University, Budapest, Hungary; Department of Functional Pharmacology, Institute of Molecular Pharmacology, Research Centre of Natural Sciences, Hungarian Academy of Sciences, Budapest, Hungary.