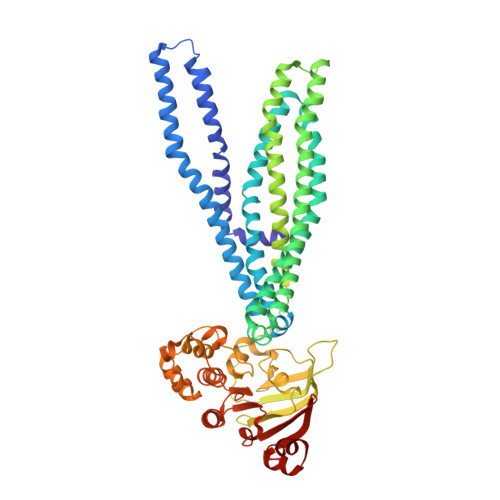
The Cystic Fibrosis Transmembrane Conductance Regulator (Cftr):3D Structure and Localisation of a Channel Gate.
Rosenberg, M.F., O'Ryan, L., Hughes, G., Zhao, Z., Aleksandrov, L.A., Riordan, J.R., Ford, R.C.(2011) J Biol Chem 286: 42647
- PubMed: 21931164
- DOI: https://doi.org/10.1074/jbc.M111.292268
- Primary Citation of Related Structures:
4A82 - PubMed Abstract:
Cystic fibrosis affects about 1 in 2500 live births and involves loss of transmembrane chloride flux due to a lack of a membrane protein channel termed the cystic fibrosis transmembrane conductance regulator (CFTR). We have studied CFTR structure by electron crystallography. The data were compared with existing structures of other ATP-binding cassette transporters. The protein was crystallized in the outward facing state and resembled the well characterized Sav1866 transporter. We identified regions in the CFTR map, not accounted for by Sav1866, which were potential locations for the regulatory region as well as the channel gate. In this analysis, we were aided by the fact that the unit cell was composed of two molecules not related by crystallographic symmetry. We also identified regions in the fitted Sav1866 model that were missing from the map, hence regions that were either disordered in CFTR or differently organized compared with Sav1866. Apart from the N and C termini, this indicated that in CFTR, the cytoplasmic end of transmembrane helix 5/11 and its associated loop could be partly disordered (or alternatively located).
Organizational Affiliation:
Faculty of Life Sciences, University of Manchester, Manchester Interdisciplinary Biocentre, Manchester M1 7DN, United Kingdom.