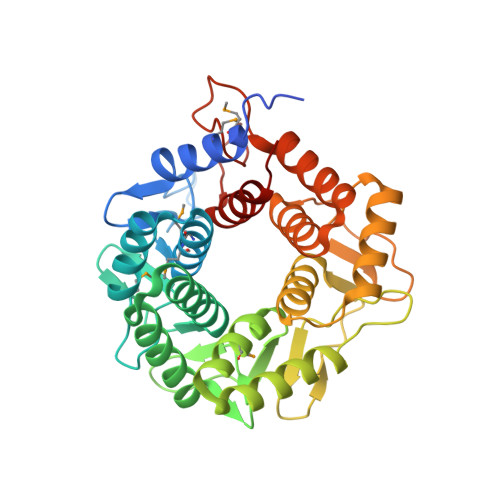
Structural analysis of CPF_2247, a novel alpha-amylase from Clostridium perfringens.
Ficko-Blean, E., Stuart, C.P., Boraston, A.B.(2011) Proteins 79: 2771-2777
- PubMed: 21905105
- DOI: https://doi.org/10.1002/prot.23116
- PubMed Abstract:
CPF_2247 from Clostridium perfringens ATCC 13124 was identified as a putative carbohydrate-active enzyme by its low sequence identity to endo-β-1,4-glucanases belonging to family 8 of the glycoside hydrolase classification. The X-ray crystal structure of CPF_2247 determined to 2.0 Å resolution by single-wavelength anomalous dispersion using seleno-methionine-substituted protein revealed an (α/α)(6) barrel fold. A large cleft on the surface of the protein contains residues that are structurally conserved with key elements of the catalytic machinery in clan GH-M glycoside hydrolases. Assessment of CPF_2247 as a carbohydrate-active enzyme disclosed α-glucanase activity on amylose, glycogen, and malto-oligosaccharides.
Organizational Affiliation:
Department of Biochemistry & Microbiology, University of Victoria, Victoria, British Columbia, Canada.