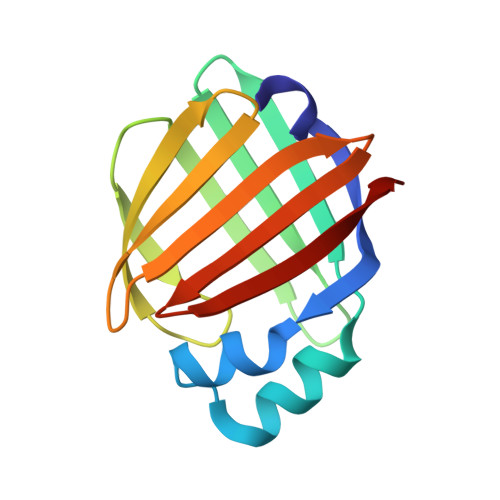
Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Cousido-Siah, A., Ayoub, D., Berberian, G., Bollo, M., Van Dorsselaer, A., Debaene, F., DiPolo, R., Petrova, T., Schulze-Briese, C., Olieric, V., Esteves, A., Mitschler, A., Sanglier-Cianferani, S., Beauge, L., Podjarny, A.(2012) Acta Crystallogr D Biol Crystallogr 68: 1098-1107
- PubMed: 22948910
- DOI: https://doi.org/10.1107/S090744491202094X
- Primary Citation of Related Structures:
3PP6, 3PPT - PubMed Abstract:
The protein ReP1-NCXSQ was isolated from the cytosol of squid nerves and has been shown to be required for MgATP stimulation of the squid nerve Na(+)/Ca(2+) exchanger NCXSQ1. In order to determine its mode of action and the corresponding biologically active ligand, sequence analysis, crystal structures and mass-spectrometric studies of this protein and its Tyr128Phe mutant are reported. Sequence analysis suggests that it belongs to the CRABP family in the FABP superfamily. The X-ray structure at 1.28 Å resolution shows the FABP β-barrel fold, with a fatty acid inside the barrel that makes a relatively short hydrogen bond to Tyr128 and shows a double bond between C9 and C10 but that is disordered beyond C12. Mass-spectrometric studies identified this fatty acid as palmitoleic acid, confirming the double bond between C9 and C10 and establishing a length of 16 C atoms in the aliphatic chain. This acid was caught inside during the culture in Escherichia coli and therefore is not necessarily linked to the biological activity. The Tyr128Phe mutant was unable to activate the Na(+)/Ca(2+) exchanger and the corresponding crystal structure showed that without the hydrogen bond to Tyr128 the palmitoleic acid inside the barrel becomes disordered. Native mass-spectrometric analysis confirmed a lower occupancy of the fatty acid in the Tyr128Phe mutant. The correlation between (i) the lack of activity of the Tyr128Phe mutant, (ii) the lower occupancy/disorder of the bound palmitoleic acid and (iii) the mass-spectrometric studies of ReP1-NCXSQ suggests that the transport of a fatty acid is involved in regulation of the NCXSQ1 exchanger, providing a novel insight into the mechanism of its regulation. In order to identify the biologically active ligand, additional high-resolution mass-spectrometric studies of the ligands bound to ReP1-NCXSQ were performed after incubation with squid nerve vesicles both with and without MgATP. These studies clearly identified palmitic acid as the fatty acid involved in regulation of the Na(+)/Ca(2+) exchanger from squid nerve.
Organizational Affiliation:
Department of Structural Biology and Genomics, IGBMC, CNRS, INSERM, Université de Strasbourg, Illkirch, France.