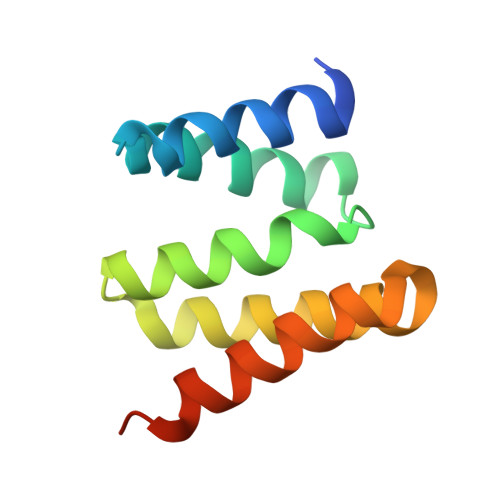
Crystal structure of the tetratricopeptide repeat domain protein Q2S6C5_SALRD from Salinibacter ruber.
Vorobiev, S., Neely, H., Seetharaman, J., Wang, H., Foote, E.L., Ciccosanti, C., Mao, L., Xiao, R., Acton, T.B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.