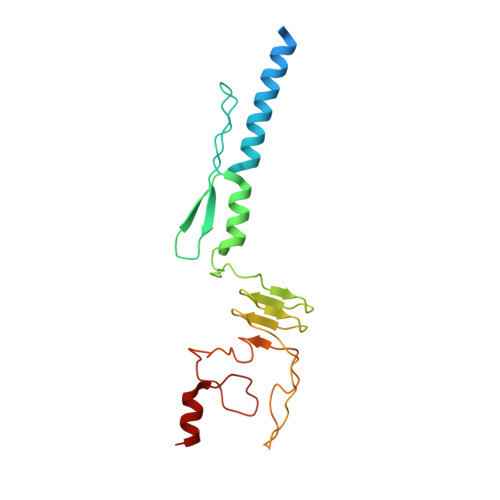
Structure of a Burkholderia pseudomallei trimeric autotransporter adhesin head.
Edwards, T.E., Phan, I., Abendroth, J., Dieterich, S.H., Masoudi, A., Guo, W., Hewitt, S.N., Kelley, A., Leibly, D., Brittnacher, M.J., Staker, B.L., Miller, S.I., Van Voorhis, W.C., Myler, P.J., Stewart, L.J.(2010) PLoS One 5: 12803-12811
- PubMed: 20862217
- DOI: https://doi.org/10.1371/journal.pone.0012803
- Primary Citation of Related Structures:
3LA9, 3LAA - PubMed Abstract:
Pathogenic bacteria adhere to the host cell surface using a family of outer membrane proteins called Trimeric Autotransporter Adhesins (TAAs). Although TAAs are highly divergent in sequence and domain structure, they are all conceptually comprised of a C-terminal membrane anchoring domain and an N-terminal passenger domain. Passenger domains consist of a secretion sequence, a head region that facilitates binding to the host cell surface, and a stalk region.
Organizational Affiliation:
Seattle Structural Genomics Center for Infectious Disease, Seattle, Washington, USA. tedwards@embios.com