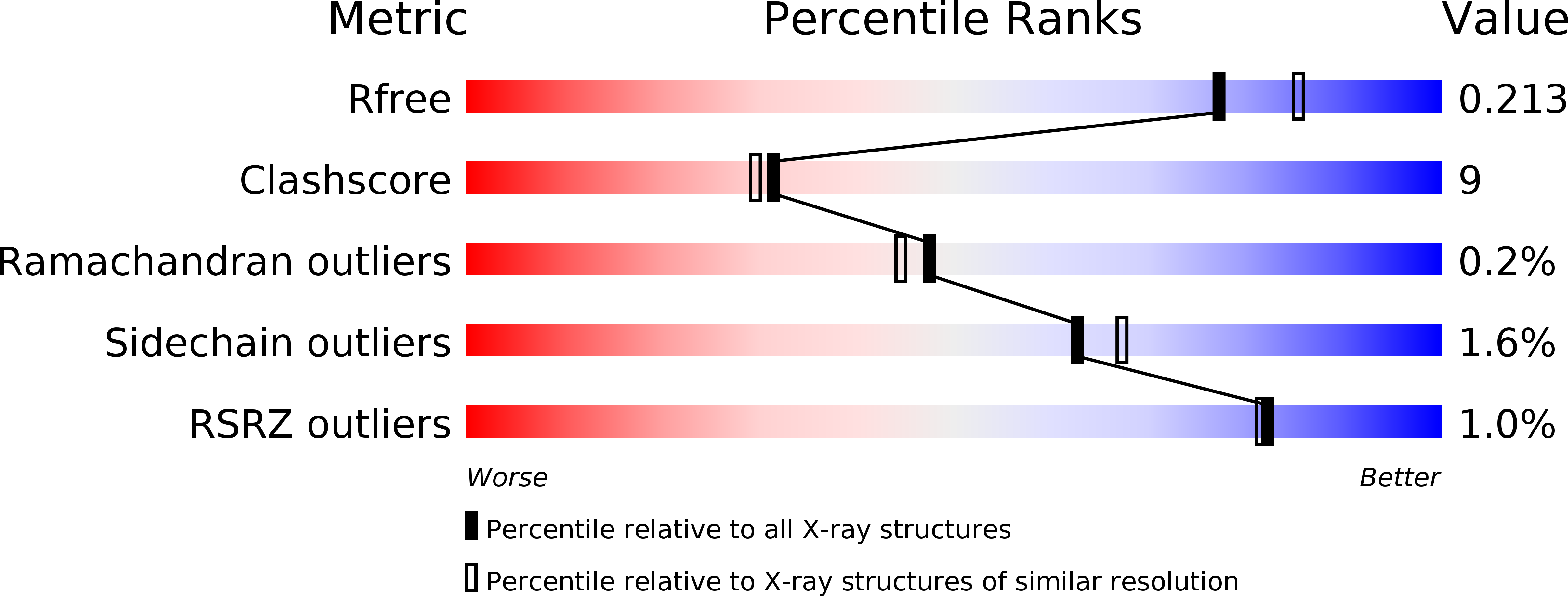
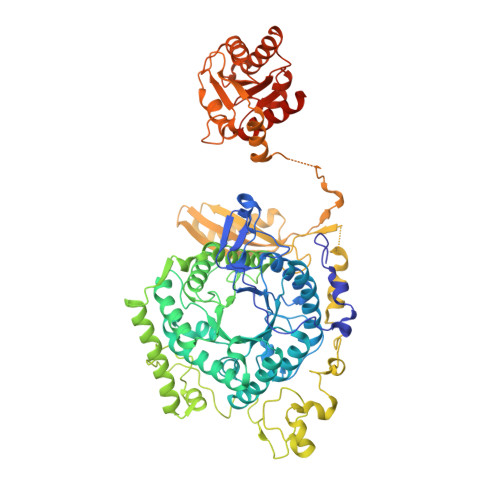
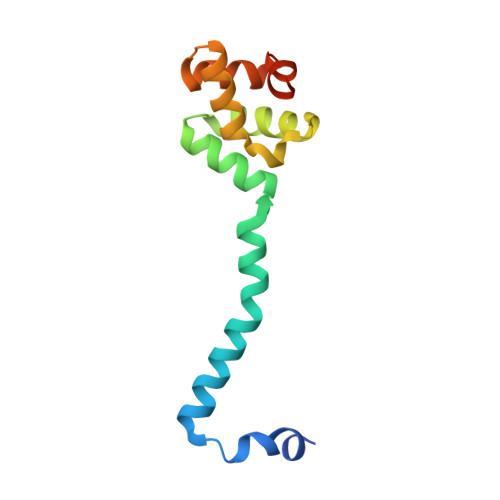
Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
Wolthers, K.R., Levy, C., Scrutton, N.S., Leys, D.(2010) J Biol Chem 285: 13942-13950
- PubMed: 20106986
- DOI: https://doi.org/10.1074/jbc.M109.068908
- Primary Citation of Related Structures:
3KOW, 3KOX, 3KOY, 3KOZ, 3KP0, 3KP1 - PubMed Abstract:
D-ornithine 4,5-aminomutase (OAM) from Clostridium sticklandii converts D-ornithine to 2,4-diaminopentanoic acid by way of radical propagation from an adenosylcobalamin (AdoCbl) to a pyridoxal 5'-phosphate (PLP) cofactor. We have solved OAM crystal structures in different catalytic states that together demonstrate unusual stability of the AdoCbl Co-C bond and that radical catalysis is coupled to large-scale domain motion. The 2.0-A substrate-free enzyme crystal structure reveals the Rossmann domain, harboring the intact AdoCbl cofactor, is tilted toward the edge of the PLP binding triose-phosphate isomerase barrel domain. The PLP forms an internal aldimine link to the Rossmann domain through Lys(629), effectively locking the enzyme in this "open" pre-catalytic conformation. The distance between PLP and 5'-deoxyadenosyl group is 23 A, and large-scale domain movement is thus required prior to radical catalysis. The OAM crystals contain two Rossmann domains within the asymmetric unit that are unconstrained by the crystal lattice. Surprisingly, the binding of various ligands to OAM crystals (in an oxygen-free environment) leads to transimination in the absence of significant reorientation of the Rossmann domains. In contrast, when performed under aerobic conditions, this leads to extreme disorder in the latter domains correlated with the loss of the 5'-deoxyadenosyl group. Our data indicate turnover and hence formation of the "closed" conformation is occurring within OAM crystals, but that the equilibrium is poised toward the open conformation. We propose that substrate binding induces large-scale domain motion concomitant with a reconfiguration of the 5'-deoxyadenosyl group, triggering radical catalysis in OAM.
Organizational Affiliation:
Faculty of Life Sciences, University of Manchester, Manchester Interdisciplinary Biocentre, 131 Princess Street, Manchester M1 7DN, United Kingdom. kirsten.wolthers@manchester.ac.uk