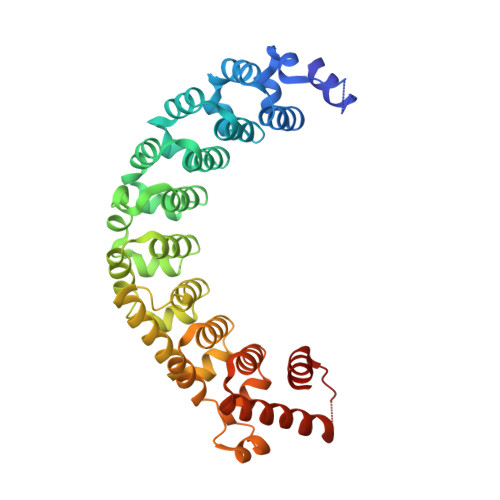
A 5' cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs.
Zhu, D., Stumpf, C.R., Krahn, J.M., Wickens, M., Hall, T.M.(2009) Proc Natl Acad Sci U S A 106: 20192-20197
- PubMed: 19918084
- DOI: https://doi.org/10.1073/pnas.0812079106
- Primary Citation of Related Structures:
3K49, 3K4E - PubMed Abstract:
A single regulatory protein can control the fate of many mRNAs with related functions. The Puf3 protein of Saccharomyces cerevisiae is exemplary, as it binds and regulates more than 100 mRNAs that encode proteins with mitochondrial function. Here we elucidate the structural basis of that specificity. To do so, we explore the crystal structures of Puf3p complexes with 2 cognate RNAs. The key determinant of Puf3p specificity is an unusual interaction between a distinctive pocket of the protein with an RNA base outside the "core" PUF-binding site. That interaction dramatically affects binding affinity in vitro and is required for regulation in vivo. The Puf3p structures, combined with those of Puf4p in the same organism, illuminate the structural basis of natural PUF-RNA networks. Yeast Puf3p binds its own RNAs because they possess a -2C and is excluded from those of Puf4p which contain an additional nucleotide in the core-binding site.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, Research Triangle Park, NC 27709, USA.