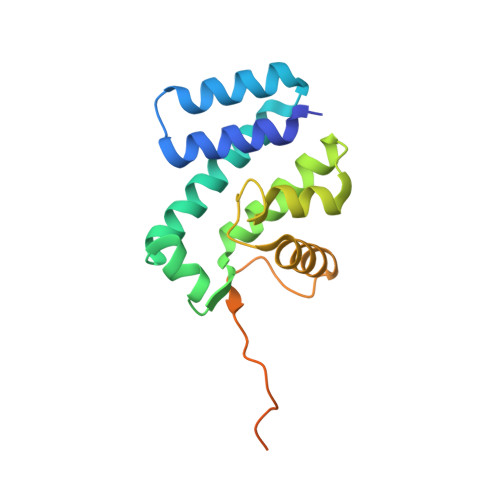
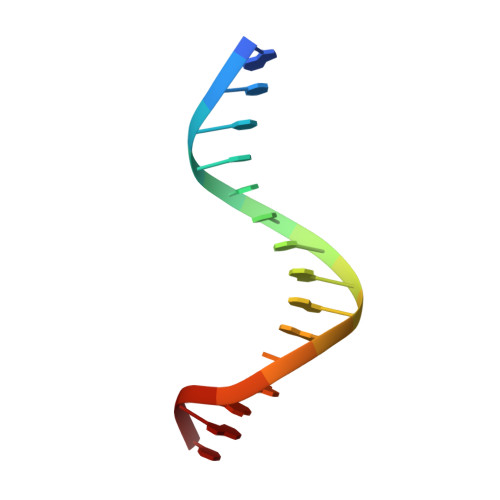
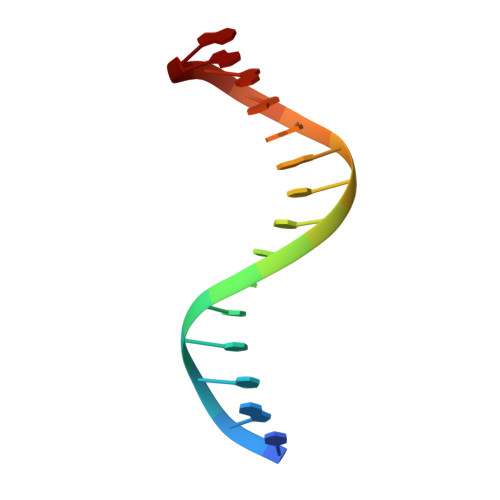
Recognition of AT-Rich DNA Binding Sites by the MogR Repressor.
Shen, A., Higgins, D.E., Panne, D.(2009) Structure 17: 769-777
- PubMed: 19446532
- DOI: https://doi.org/10.1016/j.str.2009.02.018
- Primary Citation of Related Structures:
3FDQ - PubMed Abstract:
The MogR transcriptional repressor of the intracellular pathogen Listeria monocytogenes recognizes AT-rich binding sites in promoters of flagellar genes to downregulate flagellar gene expression during infection. We describe here the 1.8 A resolution crystal structure of MogR bound to the recognition sequence 5' ATTTTTTAAAAAAAT 3' present within the flaA promoter region. Our structure shows that MogR binds as a dimer. Each half-site is recognized in the major groove by a helix-turn-helix motif and in the minor groove by a loop from the symmetry-related molecule, resulting in a "crossover" binding mode. This oversampling through minor groove interactions is important for specificity. The MogR binding site has structural features of A-tract DNA and is bent by approximately 52 degrees away from the dimer. The structure explains how MogR achieves binding specificity in the AT-rich genome of L. monocytogenes and explains the evolutionary conservation of A-tract sequence elements within promoter regions of MogR-regulated flagellar genes.
Organizational Affiliation:
Department of Microbiology and Molecular Genetics, Harvard Medical School, Boston, MA 02115, USA.