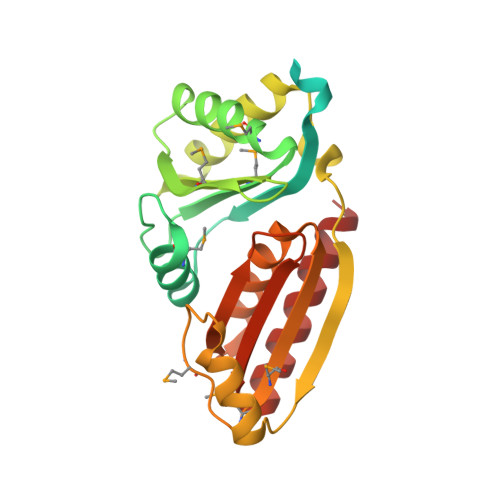
Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport.
Klein, M.G., Zwart, P., Bagby, S.C., Cai, F., Chisholm, S.W., Heinhorst, S., Cannon, G.C., Kerfeld, C.A.(2009) J Mol Biol 392: 319-333
- PubMed: 19328811
- DOI: https://doi.org/10.1016/j.jmb.2009.03.056
- Primary Citation of Related Structures:
3F56, 3FCH - PubMed Abstract:
Bacterial microcompartments (BMCs) are polyhedral bodies, composed entirely of proteins, that function as organelles in bacteria; they promote subcellular processes by encapsulating and co-localizing targeted enzymes with their substrates. The best-characterized BMC is the carboxysome, a central part of the carbon-concentrating mechanism that greatly enhances carbon fixation in cyanobacteria and some chemoautotrophs. Here we report the first structural insights into the carboxysome of Prochlorococcus, the numerically dominant cyanobacterium in the world's oligotrophic oceans. Bioinformatic methods, substantiated by analysis of gene expression data, were used to identify a new carboxysome shell component, CsoS1D, in the genome of Prochlorococcus strain MED4; orthologs were subsequently found in all cyanobacteria. Two independent crystal structures of Prochlorococcus MED4 CsoS1D reveal three features not seen in any BMC-domain protein structure solved to date. First, CsoS1D is composed of a fused pair of BMC domains. Second, this double-domain protein trimerizes to form a novel pseudohexameric building block for incorporation into the carboxysome shell, and the trimers further dimerize, forming a two-tiered shell building block. Third, and most strikingly, the large pore formed at the 3-fold axis of symmetry appears to be gated. Each dimer of trimers contains one trimer with an open pore and one whose pore is obstructed due to side-chain conformations of two residues that are invariant among all CsoS1D orthologs. This is the first evidence of the potential for gated transport across the carboxysome shell and reveals a new type of building block for BMC shells.
Organizational Affiliation:
US Department of Energy Joint Genome Institute, Walnut Creek, CA 94598, USA.