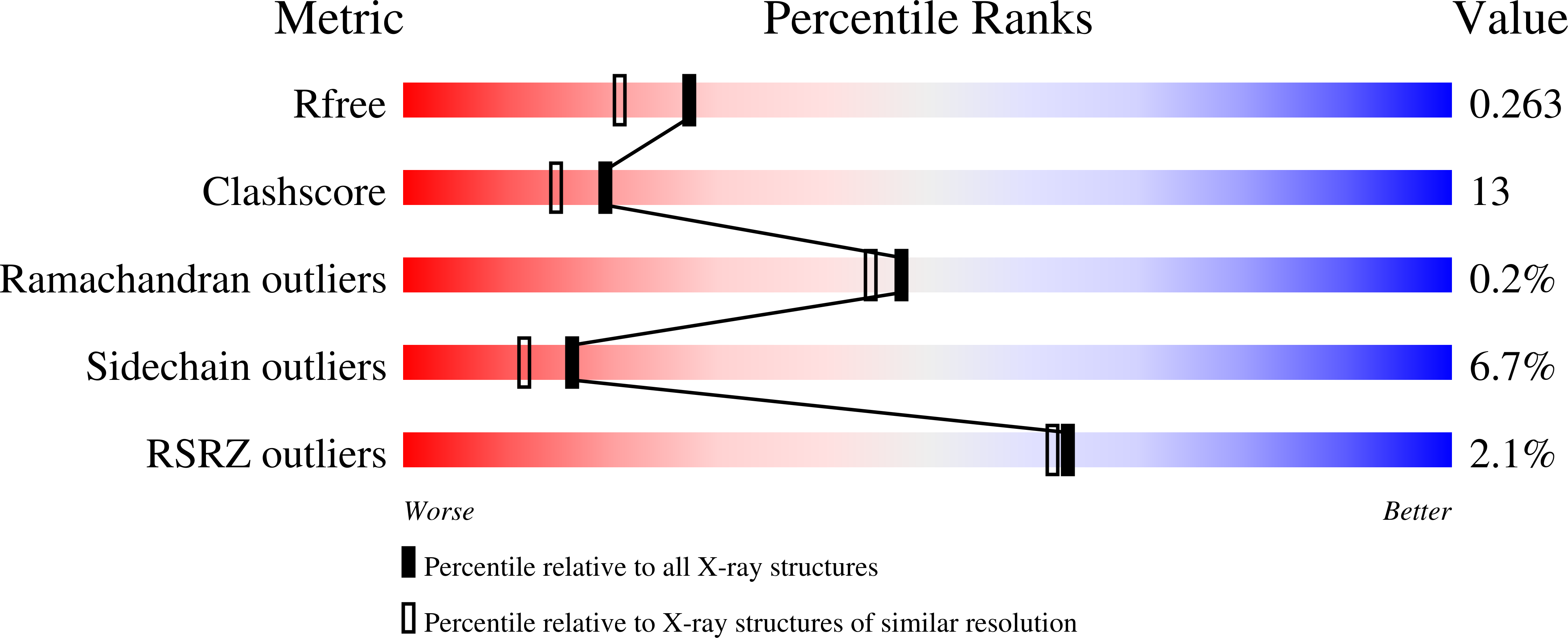
Identification of Formyl Kynurenine Formamidase and Kynurenine Aminotransferase from Saccharomyces cerevisiae Using Crystallographic, Bioinformatic and Biochemical Evidence.
Wogulis, M., Chew, E.R., Donohoue, P.D., Wilson, D.K.(2008) Biochemistry 47: 1608-1621
- PubMed: 18205391
- DOI: https://doi.org/10.1021/bi701172v
- Primary Citation of Related Structures:
3B46 - PubMed Abstract:
The essential enzymatic cofactor NAD+ can be synthesized in many eukaryotes, including Saccharomyces cerevisiae and mammals, using tryptophan as a starting material. Metabolites along the pathway or on branches have important biological functions. For example, kynurenic acid can act as an NMDA antagonist, thereby functioning as a neuroprotectant in a wide range of pathological states. N-Formyl kynurenine formamidase (FKF) catalyzes the second step of the NAD+ biosynthetic pathway by hydrolyzing N-formyl kynurenine to produce kynurenine and formate. The S. cerevisiae FKF had been reported to be a pyridoxal phosphate-dependent enzyme encoded by BNA3. We used combined crystallographic, bioinformatic and biochemical methods to demonstrate that Bna3p is not an FKF but rather is most likely the yeast kynurenine aminotransferase, which converts kynurenine to kynurenic acid. Additionally, we identify YDR428C, a yeast ORF coding for an alpha/beta hydrolase with no previously assigned function, as the FKF. We predicted its function based on our interpretation of prior structural genomics results and on its sequence homology to known FKFs. Biochemical, bioinformatics, genetic and in vivo metabolite data derived from LC-MS demonstrate that YDR428C, which we have designated BNA7, is the yeast FKF.
Organizational Affiliation:
Section of Molecular and Cellular Biology, University of California, Davis, California 95616, USA.