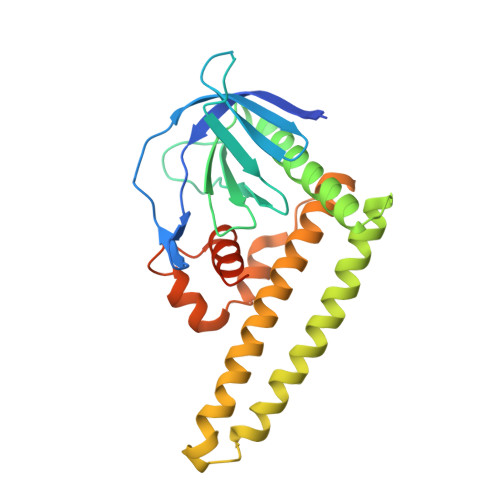
The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Terawaki, S., Kitano, K., Mori, T., Zhai, Y., Higuchi, Y., Itoh, N., Watanabe, T., Kaibuchi, K., Hakoshima, T.(2010) EMBO J 29: 236-250
- PubMed: 19893486
- DOI: https://doi.org/10.1038/emboj.2009.323
- Primary Citation of Related Structures:
3A8N, 3A8P, 3A8Q - PubMed Abstract:
Tiam1 and Tiam2 (Tiam1/2) are guanine nucleotide-exchange factors that possess the PH-CC-Ex (pleckstrin homology, coiled coil and extra) region that mediates binding to plasma membranes and signalling proteins in the activation of Rac GTPases. Crystal structures of the PH-CC-Ex regions revealed a single globular domain, PHCCEx domain, comprising a conventional PH subdomain associated with an antiparallel coiled coil of CC subdomain and a novel three-helical globular Ex subdomain. The PH subdomain resembles the beta-spectrin PH domain, suggesting non-canonical phosphatidylinositol binding. Mutational and binding studies indicated that CC and Ex subdomains form a positively charged surface for protein binding. We identified two unique acidic sequence motifs in Tiam1/2-interacting proteins for binding to PHCCEx domain, Motif-I in CD44 and ephrinB's and the NMDA receptor, and Motif-II in Par3 and JIP2. Our results suggest the molecular basis by which the Tiam1/2 PHCCEx domain facilitates dual binding to membranes and signalling proteins.
Organizational Affiliation:
Structural Biology Laboratory, Nara Institute of Science and Technology, Takayama, Ikoma, Nara, Japan.