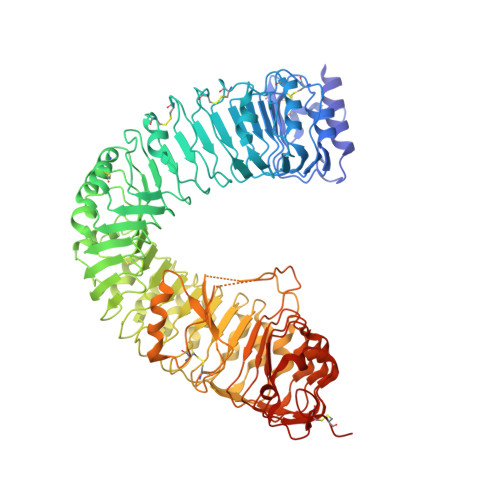
Structural insight into brassinosteroid perception by BRI1.
She, J., Han, Z., Kim, T.W., Wang, J., Cheng, W., Chang, J., Shi, S., Wang, J., Yang, M., Wang, Z.Y., Chai, J.(2011) Nature 474: 472-476
- PubMed: 21666666
- DOI: https://doi.org/10.1038/nature10178
- Primary Citation of Related Structures:
3RGX, 3RGZ - PubMed Abstract:
Brassinosteroids are essential phytohormones that have crucial roles in plant growth and development. Perception of brassinosteroids requires an active complex of BRASSINOSTEROID-INSENSITIVE 1 (BRI1) and BRI1-ASSOCIATED KINASE 1 (BAK1). Recognized by the extracellular leucine-rich repeat (LRR) domain of BRI1, brassinosteroids induce a phosphorylation-mediated cascade to regulate gene expression. Here we present the crystal structures of BRI1(LRR) in free and brassinolide-bound forms. BRI1(LRR) exists as a monomer in crystals and solution independent of brassinolide. It comprises a helical solenoid structure that accommodates a separate insertion domain at its concave surface. Sandwiched between them, brassinolide binds to a hydrophobicity-dominating surface groove on BRI1(LRR). Brassinolide recognition by BRI1(LRR) is through an induced-fit mechanism involving stabilization of two interdomain loops that creates a pronounced non-polar surface groove for the hormone binding. Together, our results define the molecular mechanisms by which BRI1 recognizes brassinosteroids and provide insight into brassinosteroid-induced BRI1 activation.
Organizational Affiliation:
Key Laboratory for Protein Sciences of Ministry of Education School of Life Sciences, Tsinghua University, Beijing 100084, China.