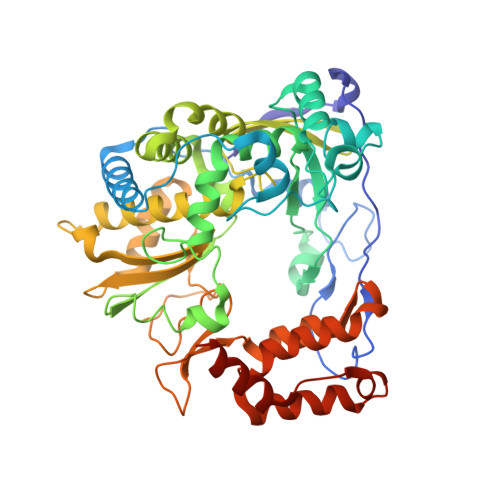
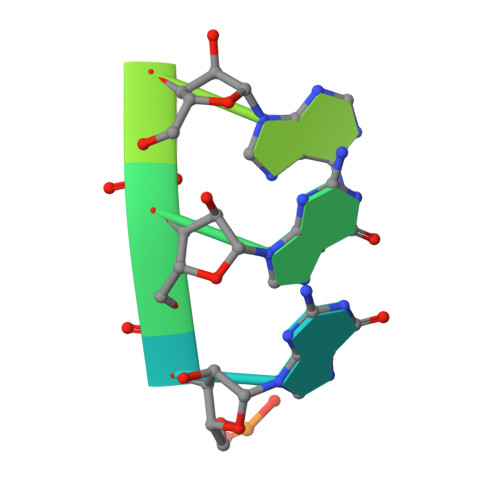
Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase.
Gong, P., Peersen, O.B.(2010) Proc Natl Acad Sci U S A 107: 22505-22510
- PubMed: 21148772
- DOI: https://doi.org/10.1073/pnas.1007626107
- Primary Citation of Related Structures:
3OL6, 3OL7, 3OL8, 3OL9, 3OLA, 3OLB - PubMed Abstract:
Positive-strand RNA viruses include a large number of human and animal pathogens whose essential RNA-dependent RNA polymerases (RdRPs) share a structurally homologous core with an encircled active site. RdRPs are targets for antiviral drug development, but these efforts are hindered by limited structural information about the RdRP catalytic cycle. To further our understanding of RdRP function, we assembled, purified, and then crystallized poliovirus elongation complexes after multiple rounds of nucleotide incorporation. Here we present structures capturing the active polymerase and its nucleotide triphosphate complexes in four distinct states, leading us to propose a six-state catalytic cycle involving residues that are highly conserved among positive-strand RNA virus RdRPs. The structures indicate that RdRPs use a fully prepositioned templating base for nucleotide recognition and close their active sites for catalysis using a novel structural rearrangement in the palm domain. The data also suggest that translocation by RDRPs may not be directly linked to the conformational changes responsible for active site closure and reopening.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Colorado State University, Fort Collins, CO 80523-1870, USA.