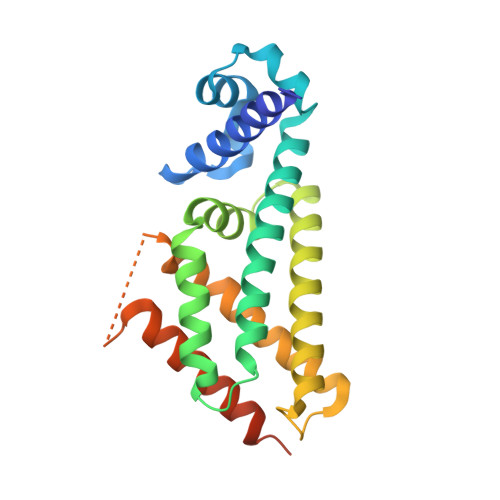
Structural basis for the transcriptional regulation of membrane lipid homeostasis.
Miller, D.J., Zhang, Y.M., Subramanian, C., Rock, C.O., White, S.W.(2010) Nat Struct Mol Biol 17: 971-975
- PubMed: 20639888
- DOI: https://doi.org/10.1038/nsmb.1847
- Primary Citation of Related Structures:
3LSJ, 3LSP, 3LSR - PubMed Abstract:
DesT is a transcriptional repressor that regulates the genes that control the unsaturated:saturated fatty acid ratio available for membrane lipid synthesis. DesT bound to unsaturated acyl-CoA has a high affinity for its cognate palindromic DNA-binding site, whereas DesT bound to saturated acyl-CoA does not bind this site. Structural analyses of the DesT-oleoyl-CoA-DNA and DesT-palmitoyl-CoA complexes reveal that acyl chain shape directly influences the packing of hydrophobic core residues within the DesT ligand-binding domain. These changes are propagated to the paired DNA-binding domains via conformational changes to modulate DNA binding. These structural interpretations are supported by the in vitro and in vivo characterization of site-directed mutants. The regulation of DesT by the unsaturated:saturated ratio of acyl chains rather than the concentration of a single ligand is a paradigm for understanding transcriptional regulation of membrane lipid homeostasis.
Organizational Affiliation:
Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, Tennessee, USA.