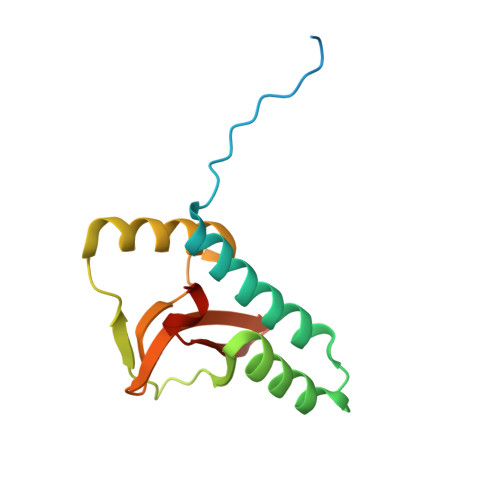
Structural basis for pH sensing by the periplsamic domain of the risS histidine kinase from Burkholderia pseudomallei
Edwards, T.E., Abendroth, J., Staker, B.L., Phan, I., Myler, P., Stacy, R., Stewart, L., Miller, S.I., Seattle Structural Genomics Center for Infectious Disease (SSGCID)To be published.