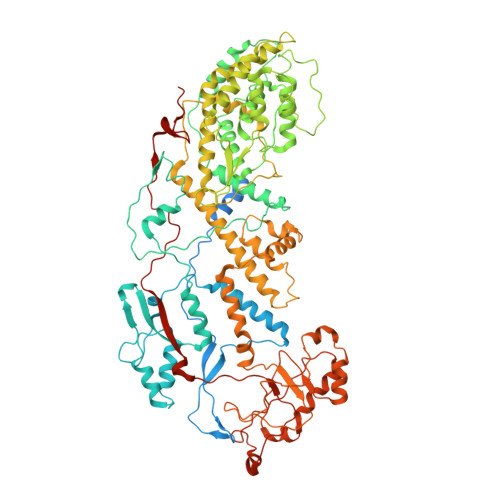
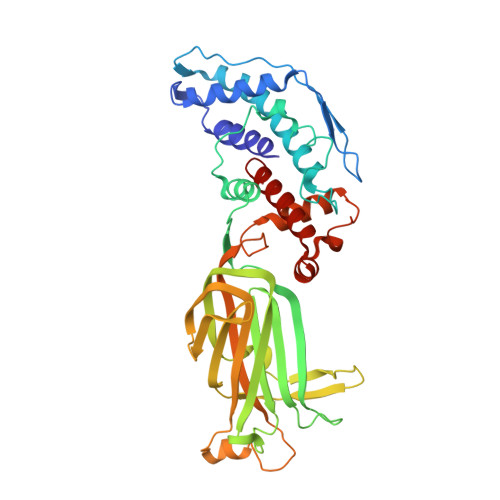
X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution.
McClain, B., Settembre, E., Temple, B.R., Bellamy, A.R., Harrison, S.C.(2010) J Mol Biol 397: 587-599
- PubMed: 20122940
- DOI: https://doi.org/10.1016/j.jmb.2010.01.055
- Primary Citation of Related Structures:
3KZ4 - PubMed Abstract:
The rotavirus inner capsid particle, known as the "double-layered particle" (DLP), is the "payload" delivered into a cell in the process of viral infection. Its inner and outer protein layers, composed of viral protein (VP) 2 and VP6, respectively, package the 11 segments of the double-stranded RNA (dsRNA) of the viral genome, as well as about the same number of polymerase molecules (VP1) and capping-enzyme molecules (VP3). We have determined the crystal structure of the bovine rotavirus DLP. There is one full particle (outer diameter approximately 700 A) in the asymmetric unit of the P2(1)2(1)2(1) unit cell of dimensions a=740 A, b=1198 A, and c=1345 A. A three-dimensional reconstruction from electron cryomicroscopy was used as a molecular replacement model for initial phase determination to about 18.5 A resolution, and the 60-fold redundancy of icosahedral particle symmetry allowed phases to be extended stepwise to the limiting resolution of the data (3.8 A). The structure of a VP6 trimer (determined previously by others) fits the outer layer density with very little adjustment. The T=13 triangulation number of that layer implies that there are four and one-third VP6 trimers per icosahedral asymmetric unit. The inner layer has 120 copies of VP2 and thus 2 copies per icosahedral asymmetric unit, designated VP2A and VP2B. Residues 101-880 fold into a relatively thin principal domain, comma-like in outline, shaped such that only rather modest distortions (concentrated at two "subdomain" boundaries) allow VP2A and VP2B to form a uniform layer with essentially no gaps at the subunit boundaries, except for a modest pore along the 5-fold axis. The VP2 principal domain resembles those of the corresponding shells and homologous proteins in other dsRNA viruses: lambda1 in orthoreoviruses and VP3 in orbiviruses. Residues 1-80 of VP2A and VP2B fold together with four other such pairs into a "5-fold hub" that projects into the DLP interior along the 5-fold axis; residues 81-100 link the 10 polypeptide chains emerging from a 5-fold hub to the N-termini of their corresponding principal domains, clustered into a decameric assembly unit. The 5-fold hub appears to have several distinct functions. One function is to recruit a copy of VP1 (or of a VP1-VP3 complex), potentially along with a segment of plus-strand RNA, as a decamer of VP2 assembles. The second function is to serve as a shaft around which can coil a segment of dsRNA. The third function is to guide nascent mRNA, synthesized in the DLP interior by VP1 and 5'-capped by the action of VP3, out through a 5-fold exit channel. We propose a model for rotavirus particle assembly, based on known requirements for virion formation, together with the structure of the DLP and that of VP1, determined earlier.
Organizational Affiliation:
Laboratory of Molecular Medicine, Children's Hospital Boston, 320 Longwood Avenue, Boston, MA 02115, USA.