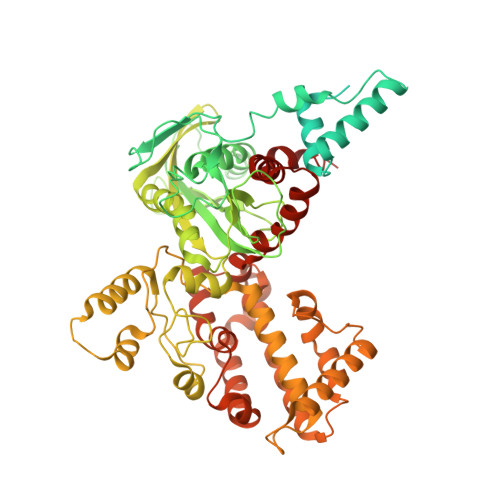
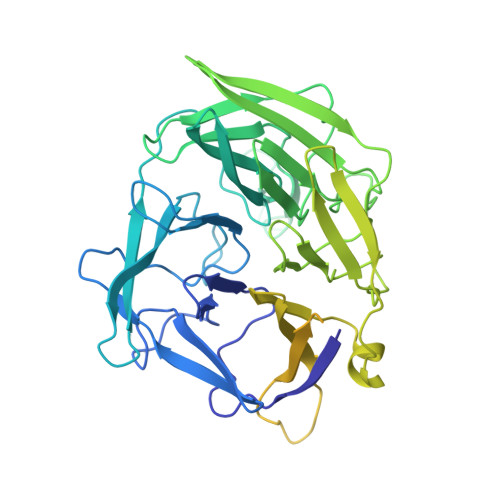
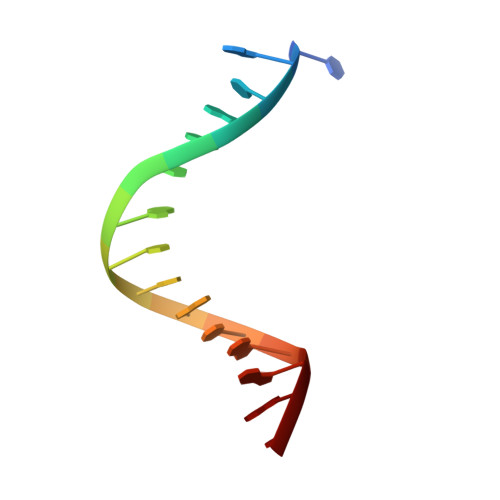
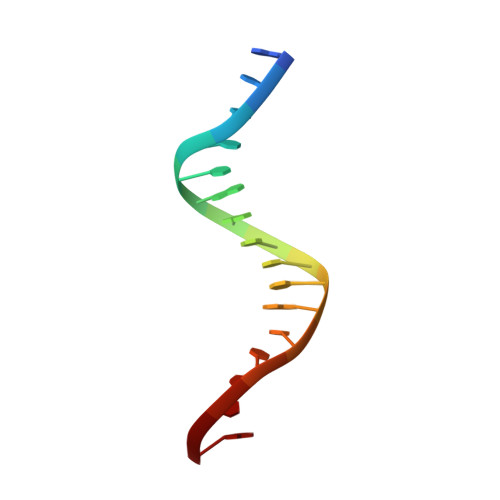
Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Ru, H., Chambers, M.G., Fu, T.M., Tong, A.B., Liao, M., Wu, H.(2015) Cell 163: 1138-1152
- PubMed: 26548953
- DOI: https://doi.org/10.1016/j.cell.2015.10.055
- Primary Citation of Related Structures:
3JBW, 3JBX, 3JBY - PubMed Abstract:
Diverse repertoires of antigen-receptor genes that result from combinatorial splicing of coding segments by V(D)J recombination are hallmarks of vertebrate immunity. The (RAG1-RAG2)2 recombinase (RAG) recognizes recombination signal sequences (RSSs) containing a heptamer, a spacer of 12 or 23 base pairs, and a nonamer (12-RSS or 23-RSS) and introduces precise breaks at RSS-coding segment junctions. RAG forms synaptic complexes only with one 12-RSS and one 23-RSS, a dogma known as the 12/23 rule that governs the recombination fidelity. We report cryo-electron microscopy structures of synaptic RAG complexes at up to 3.4 Å resolution, which reveal a closed conformation with base flipping and base-specific recognition of RSSs. Distortion at RSS-coding segment junctions and base flipping in coding segments uncover the two-metal-ion catalytic mechanism. Induced asymmetry involving tilting of the nonamer-binding domain dimer of RAG1 upon binding of HMGB1-bent 12-RSS or 23-RSS underlies the molecular mechanism for the 12/23 rule.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA; Program in Cellular and Molecular Medicine, Boston Children's Hospital, Boston, MA 02115, USA.