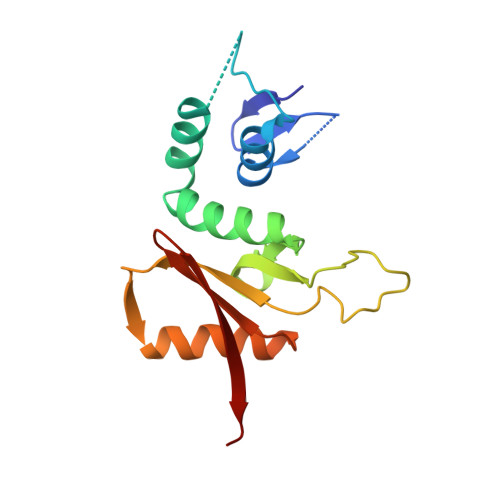
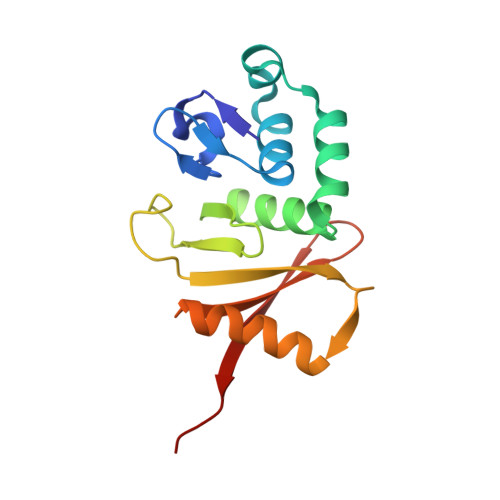
Crystal structure and assembly of the functional Nanoarchaeum equitans tRNA splicing endonuclease.
Mitchell, M., Xue, S., Erdman, R., Randau, L., Soll, D., Li, H.(2009) Nucleic Acids Res 37: 5793-5802
- PubMed: 19578064
- DOI: https://doi.org/10.1093/nar/gkp537
- Primary Citation of Related Structures:
3IEY, 3IF0 - PubMed Abstract:
The RNA splicing and processing endonuclease from Nanoarchaeum equitans (NEQ) belongs to the recently identified (alphabeta)(2) family of splicing endonucleases that require two different subunits for splicing activity. N. equitans splicing endonuclease comprises the catalytic subunit (NEQ205) and the structural subunit (NEQ261). Here, we report the crystal structure of the functional NEQ enzyme at 2.1 A containing both subunits, as well as that of the NEQ261 subunit alone at 2.2 A. The functional enzyme resembles previously known alpha(2) and alpha(4) endonucleases but forms a heterotetramer: a dimer of two heterodimers of the catalytic subunit (NEQ205) and the structural subunit (NEQ261). Surprisingly, NEQ261 alone forms a homodimer, similar to the previously known homodimer of the catalytic subunit. The homodimers of isolated subunits are inhibitory to heterodimerization as illustrated by a covalently linked catalytic homodimer that had no RNA cleavage activity upon mixing with the structural subunit. Detailed structural comparison reveals a more favorable hetero- than homodimerization interface, thereby suggesting a possible regulation mechanism of enzyme assembly through available subunits. Finally, the uniquely flexible active site of the NEQ endonuclease provides a possible explanation for its broader substrate specificity.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Institute of Molecular Biophysics, Florida State University, Tallahassee, FL 32306, USA.