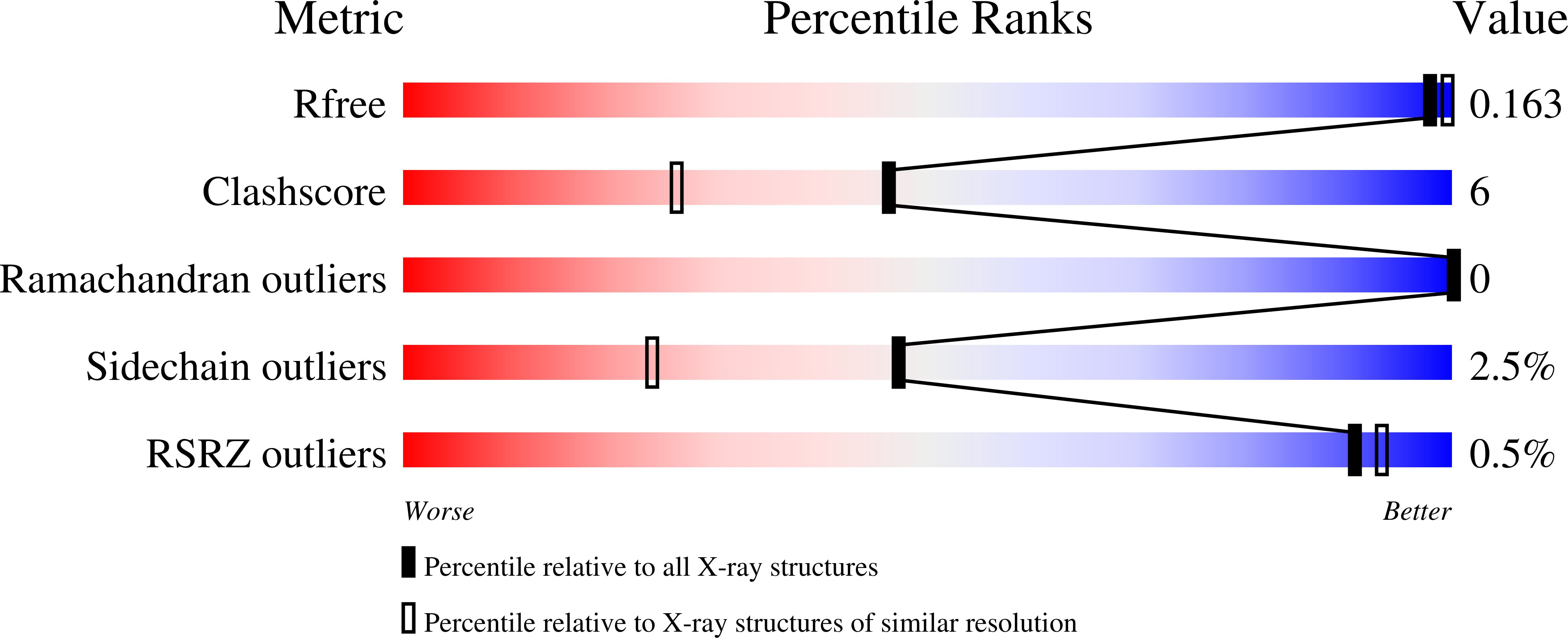
Correlation between hemichrome stability and the root effect in tetrameric hemoglobins.
Vergara, A., Franzese, M., Merlino, A., Bonomi, G., Verde, C., Giordano, D., di Prisco, G., Lee, H.C., Peisach, J., Mazzarella, L.(2009) Biophys J 97: 866-874
- PubMed: 19651045
- DOI: https://doi.org/10.1016/j.bpj.2009.04.056
- Primary Citation of Related Structures:
3GQG - PubMed Abstract:
Oxidation of Hbs leads to the formation of different forms of Fe(III) that are relevant to a range of biochemical and physiological functions. Here we report a combined EPR/x-ray crystallography study performed at acidic pH on six ferric tetrameric Hbs. Five of the Hbs were isolated from the high-Antarctic notothenioid fishes Trematomus bernacchii, Trematomus newnesi, and Gymnodraco acuticeps, and one was isolated from the sub-Antarctic notothenioid Cottoperca gobio. Our EPR analysis reveals that 1), in all of these Hbs, at acidic pH the aquomet form and two hemichromes coexist; and 2), only in the three Hbs that exhibit the Root effect is a significant amount of the pentacoordinate (5C) high-spin Fe(III) form found. The crystal structure at acidic pH of the ferric form of the Root-effect Hb from T. bernacchii is also reported at 1.7 A resolution. This structure reveals a 5C state of the heme iron for both the alpha- and beta-chains within a T quaternary structure. Altogether, the spectroscopic and crystallographic results indicate that the Root effect and hemichrome stability at acidic pH are correlated in tetrameric Hbs. Furthermore, Antarctic fish Hbs exhibit higher peroxidase activity than mammalian and temperate fish Hbs, suggesting that a partial hemichrome state in tetrameric Hbs, unlike in monomeric Hbs, does not remove the need for protection from peroxide attack, in contrast to previous results from monomeric Hbs.
Organizational Affiliation:
Department of Chemistry, University of Naples Federico II, Complesso Universitario Monte S. Angelo, Naples, Italy.