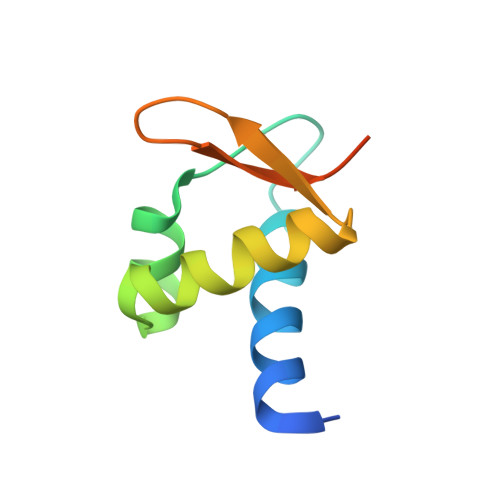
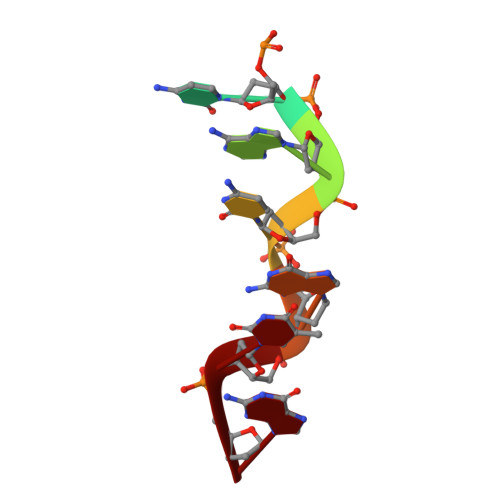The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Ha, S.C., Choi, J., Hwang, H.Y., Rich, A., Kim, Y.G., Kim, K.K.(2009) Nucleic Acids Res 37: 629-637
- PubMed: 19074195
- DOI: https://doi.org/10.1093/nar/gkn976
- Primary Citation of Related Structures:
3F21, 3F22, 3F23 - PubMed Abstract:
The Z-DNA conformation preferentially occurs at alternating purine-pyrimidine repeats, and is specifically recognized by Z alpha domains identified in several Z-DNA-binding proteins. The binding of Z alpha to foreign or chromosomal DNA in various sequence contexts is known to influence various biological functions, including the DNA-mediated innate immune response and transcriptional modulation of gene expression. For these reasons, understanding its binding mode and the conformational diversity of Z alpha bound Z-DNAs is of considerable importance. However, structural studies of Z alpha bound Z-DNA have been mostly limited to standard CG-repeat DNAs. Here, we have solved the crystal structures of three representative non-CG repeat DNAs, d(CACGTG)(2), d(CGTACG)(2) and d(CGGCCG)(2) complexed to hZ alpha(ADAR1) and compared those structures with that of hZ alpha(ADAR1)/d(CGCGCG)(2) and the Z alpha-free Z-DNAs. hZ alpha(ADAR1) bound to each of the three Z-DNAs showed a well conserved binding mode with very limited structural deviation irrespective of the DNA sequence, although varying numbers of residues were in contact with Z-DNA. Z-DNAs display less structural alterations in the Z alpha-bound state than in their free form, thereby suggesting that conformational diversities of Z-DNAs are restrained by the binding pocket of Z alpha. These data suggest that Z-DNAs are recognized by Z alpha through common conformational features regardless of the sequence and structural alterations.
Organizational Affiliation:
Department of Molecular Cell Biology, Samsung Biomedical Research Institute, Sungkyunkwan University School of Medicine, Suwon 440-746, Korea.