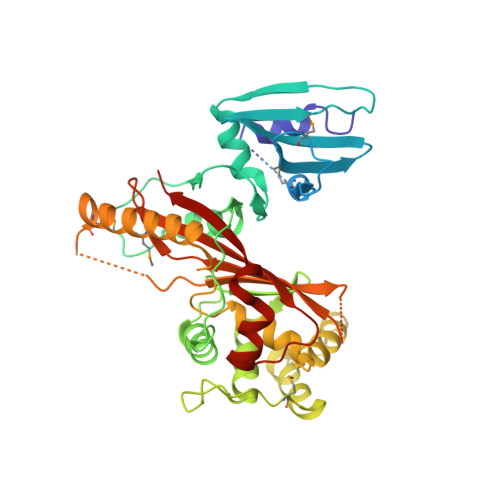
Novel fold of VirA, a type III secretion system effector protein from Shigella flexneri
Davis, J., Wang, J., Tropea, J.E., Zhang, D., Dauter, Z., Waugh, D.S., Wlodawer, A.(2008) Protein Sci 17: 2167-2173
- PubMed: 18787201
- DOI: https://doi.org/10.1110/ps.037978.108
- Primary Citation of Related Structures:
3EE1 - PubMed Abstract:
VirA, a secreted effector protein from Shigella sp., has been shown to be necessary for its virulence. It was also reported that VirA might be related to papain-like cysteine proteases and cleave alpha-tubulin, thus facilitating intracellular spreading. We have now determined the crystal structure of VirA at 3.0 A resolution. The shape of the molecule resembles the letter "V," with the residues in the N-terminal third of the 45-kDa molecule (some of which are disordered) forming one clearly identifiable domain, and the remainder of the molecule completing the V-like structure. The fold of VirA is unique and does not resemble that of any known protein, including papain, although its N-terminal domain is topologically similar to cysteine protease inhibitors such as stefin B. Analysis of the sequence conservation between VirA and its Escherichia coli homologs EspG and EspG2 did not result in identification of any putative protease-like active site, leaving open a possibility that the biological function of VirA in Shigella virulence may not involve direct proteolytic activity.
Organizational Affiliation:
Protein Structure Section, Macromolecular Crystallography Laboratory, NCI, Frederick, Maryland 21702, USA.