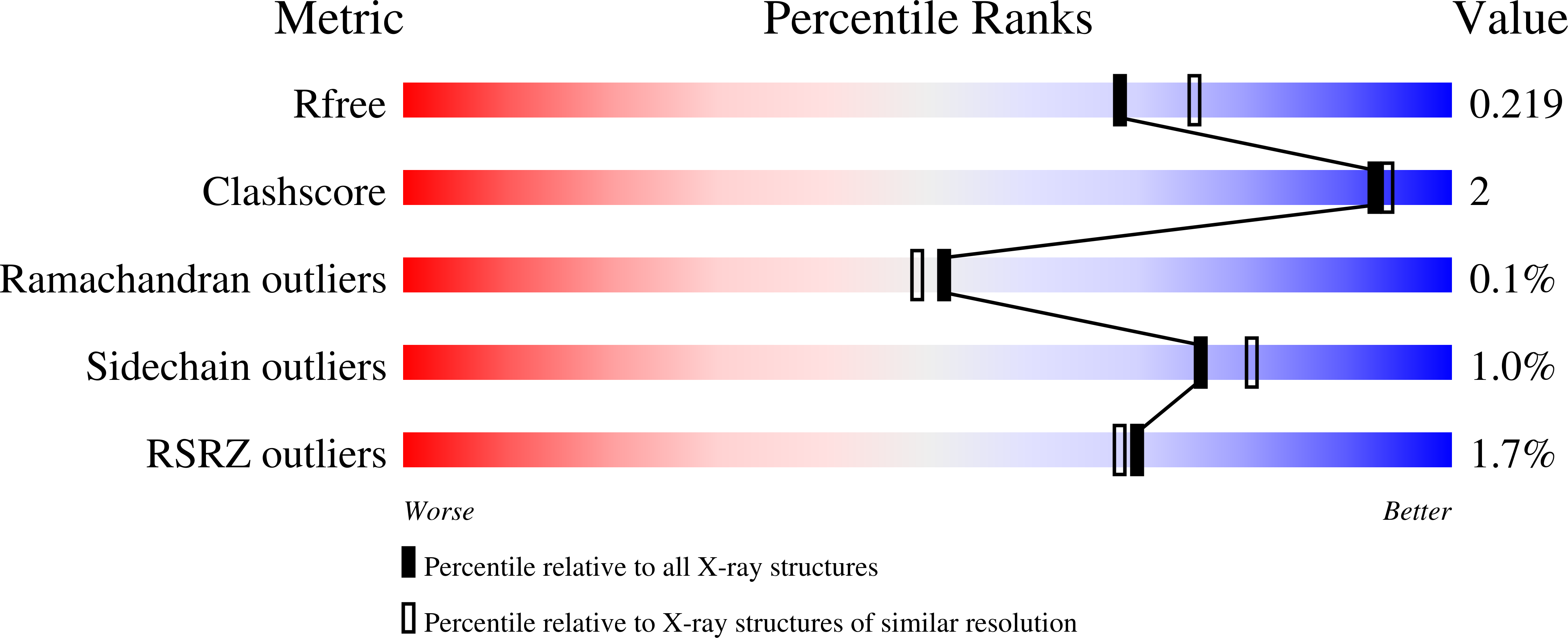
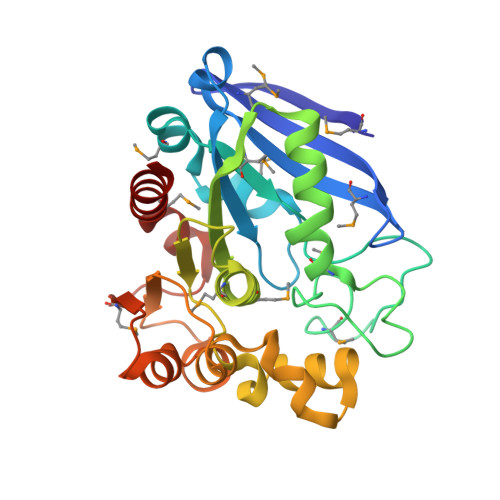
The structure of a putative S-formylglutathione hydrolase from Agrobacterium tumefaciens
van Straaten, K.E., Gonzalez, C.F., Valladares, R.B., Xu, X., Savchenko, A.V., Sanders, D.A.(2009) Protein Sci 18: 2196-2202
- PubMed: 19653299
- DOI: https://doi.org/10.1002/pro.216
- Primary Citation of Related Structures:
3E4D - PubMed Abstract:
The structure of the Atu1476 protein from Agrobacterium tumefaciens was determined at 2 A resolution. The crystal structure and biochemical characterization of this enzyme support the conclusion that this protein is an S-formylglutathione hydrolase (AtuSFGH). The three-dimensional structure of AtuSFGH contains the alpha/beta hydrolase fold topology and exists as a homo-dimer. Contacts between the two monomers in the dimer are formed both by hydrogen bonds and salt bridges. Biochemical characterization reveals that AtuSFGH hydrolyzes C--O bonds with high affinity toward short to medium chain esters, unlike the other known SFGHs which have greater affinity toward shorter chained esters. A potential role for Cys54 in regulation of enzyme activity through S-glutathionylation is also proposed.
Organizational Affiliation:
Department of Chemistry, University of Saskatchewan, Saskatoon, SK, Canada.