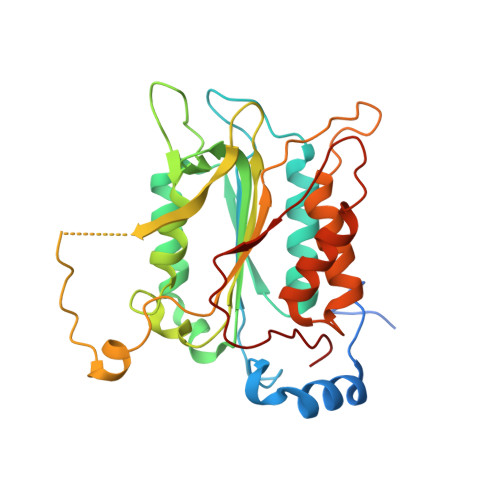
Crystal structure of procaspase-1 zymogen domain reveals insight into inflammatory caspase autoactivation
Elliott, J.M., Rouge, L., Wiesmann, C., Scheer, J.M.(2009) J Biol Chem 284: 6546-6553
- PubMed: 19117953
- DOI: https://doi.org/10.1074/jbc.M806121200
- Primary Citation of Related Structures:
3E4C - PubMed Abstract:
One key event in inflammatory signaling is the activation of the initiator caspase, procaspase-1. Presented here is the crystal structure of the procaspase-1 zymogen without its caspase recruitment domain solved to 2.05 A. Although the isolated domain is monomeric in solution, the protein appeared dimeric in crystals. The loop arrangements in the dimer provide insight into the first autoproteolytic events that occur during activation by oligomerization. Additionally, in contrast to other caspases, we demonstrate that autoproteolysis at the second cleavage site, Asp316, is necessary for conversion to a stable dimer in solution. Critical elements of secondary structure are revealed in the crystal structure that explain why a dimeric protein is favored after proteolysis at this aspartic acid. Dimer stabilization is concurrent with a 130-fold increase in kcat, the sole contributing kinetic factor to an activated and efficient mediator of inflammation.
Organizational Affiliation:
Department of Protein Chemistry, Genentech, Inc., South San Francisco, California 94080, USA.