Structure and mechanism of a cofactor f0 accelerated (6-4) photolyase from the fruit fly
Maul, M.J., Barends, T., Glas, A.F., Cryle, M.J., Schneider, S., Domratcheva, T., Schlichting, I., Carell, T.To be published.
Experimental Data Snapshot
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RE11660p | C [auth A] | 543 | Drosophila melanogaster | Mutation(s): 1 Gene Names: phr64-520 | 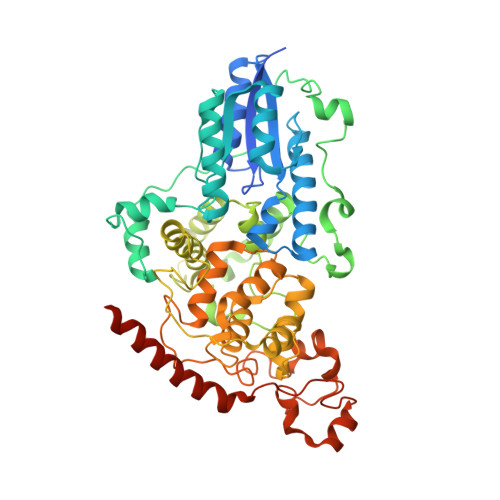 |
UniProt | |||||
Find proteins for Q8SXK5 (Drosophila melanogaster) Explore Q8SXK5 Go to UniProtKB: Q8SXK5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8SXK5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3') | A [auth C] | 15 | N/A | 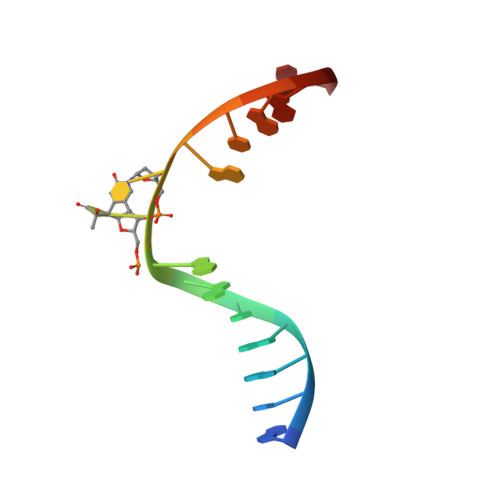 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3') | B [auth D] | 15 | N/A | 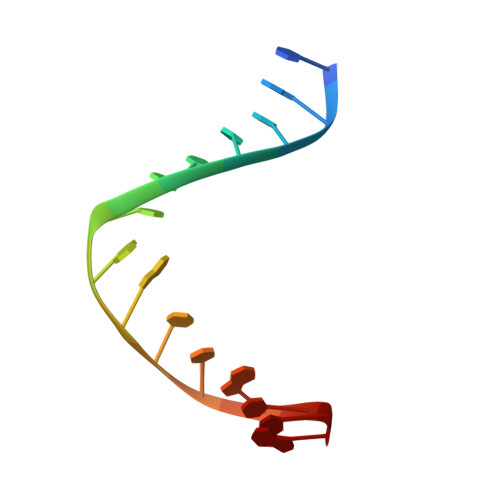 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD Query on FAD | D [auth A] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| FO1 Query on FO1 | E [auth A] | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol C16 H17 N3 O7 AUEILLWDYUBWCM-XQQFMLRXSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 86.47 | α = 90 |
| b = 89.51 | β = 90 |
| c = 89.34 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| MAR345 | data collection |
| XDS | data reduction |
| XSCALE | data scaling |
| REFMAC | phasing |