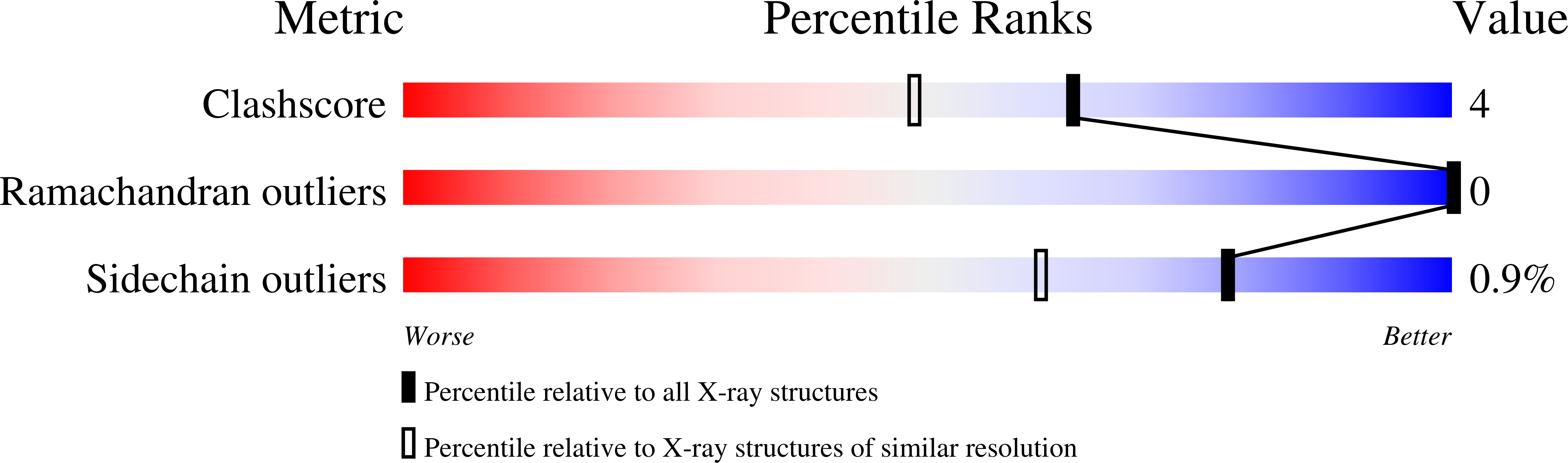Design of an engineered N-terminal HIV-1 gp41 trimer with enhanced stability and potency
Dwyer, J.J., Wilson, K.L., Martin, K., Seedorff, J.E., Hasan, A., Medinas, R.J., Davison, D.K., Feese, M.D., Richter, H.T., Kim, H., Matthews, T.J., Delmedico, M.K.(2008) Protein Sci 17: 633-643
- PubMed: 18359857
- DOI: https://doi.org/10.1110/ps.073307608
- Primary Citation of Related Structures:
2ZFC - PubMed Abstract:
HIV fusion is mediated by a conformational transition in which the C-terminal region (HR2) of gp41 interacts with the N-terminal region (HR1) to form a six-helix bundle. Peptides derived from the HR1 form a well-characterized, trimeric coiled-coil bundle in the presence of HR2 peptides, but there is little structural information on the isolated HR1 trimer. Using protein design, we have designed synthetic HR1 peptides that form soluble, thermostable HR1 trimers. In vitro binding of HR2 peptides to the engineered trimer suggests that the design strategy has not significantly impacted the ability to form the six-helix bundle. The peptides have enhanced antiviral activity compared to wild type, with up to 30-fold greater potency against certain viral isolates. In vitro passaging was used to generate HR1-resistant virus and the observed resistance mutations map to the HR2 region of gp41, demonstrating that the peptides block the fusion process by binding to the viral HR2 domain. Interestingly, the activity of the HR2 fusion inhibitor, enfuvirtide (ENF), against these resistant viruses is maintained or improved up to fivefold. The 1.5 A crystal structure of one of these designs has been determined, and we show that the isolated HR1 is very similar to the conformation of the HR1 in the six-helix bundle. These results provide an initial model of the pre-fusogenic state, are attractive starting points for identifying novel fusion inhibitors, and offer new opportunities for developing HIV therapeutics based on HR1 peptides.
Organizational Affiliation:
Trimeris, Inc., Protein Engineering Group, Morrisville, North Carolina 27560, USA. jdwyer@trimeris.com