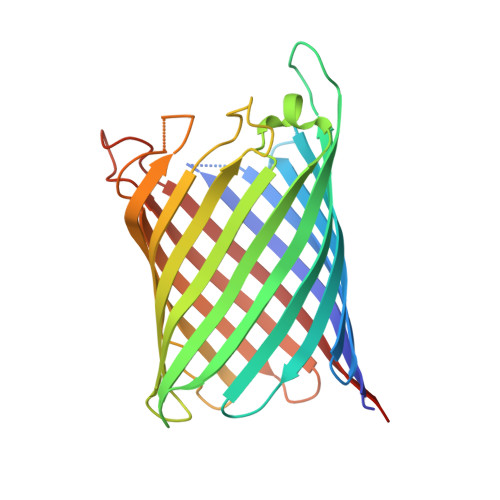
Structural and Functional Characterization of a Synthetically Modified Ompg.
Grosse, W., Reiss, P., Reitz, S., Cebi, M., Luebben, W., Koert, U., Essen, L.-O.(2010) Bioorg Med Chem 18: 7716
- PubMed: 20378361
- DOI: https://doi.org/10.1016/j.bmc.2010.03.044
- Primary Citation of Related Structures:
2WVP - PubMed Abstract:
Chemical modification of ion channels has recently attracted attention due to their potential use in stochastic sensing and neurobiology. Among the available channel templates stable β-barrel proteins have shown their potential for large scale chemical modifications due to their wide pore lumen. Ion-channel hybrids using the outer membrane protein OmpG were generated by S-alkylation with a synthetic modulator and functionally as well as structurally characterized. The dansyl moiety of the used modulator resulted in partial blockage of current though the OmpG channel with its gating characteristics mainly unaffected. The crystal structure of an OmpG-dansyl hybrid at 2.4Å resolution correlates this finding by showing that the modulator lines the inner walling of the OmpG pore. These results underline the suitability of OmpG as a structural base for the construction of stochastic sensors.
Organizational Affiliation:
Philipps-Universität Marburg, Fachbereich Chemie, Hans-Meerwein-Straße, 35032 Marburg, Germany.