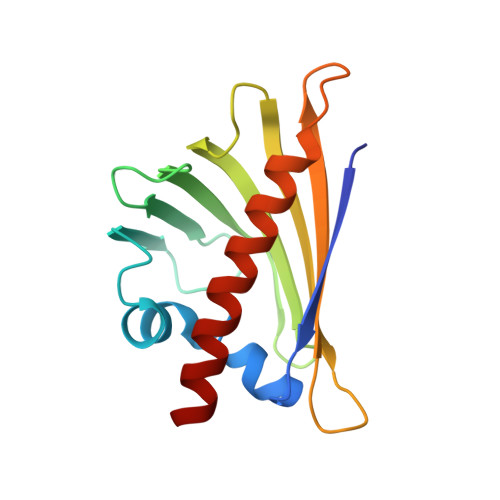
Structure of the Major Carrot Allergen Dau C 1.
Markovic-Housley, Z., Basle, A., Padavattan, S., Maderegger, B., Schirmer, T., Hoffmann-Sommergruber, K.(2009) Acta Crystallogr D Biol Crystallogr 65: 1206
- PubMed: 19923716
- DOI: https://doi.org/10.1107/S0907444909034854
- Primary Citation of Related Structures:
2WQL - PubMed Abstract:
Dau c 1 is a major allergen of carrot (Daucus carota) which displays IgE cross-reactivity with the homologous major birch-pollen allergen Bet v 1. The crystal structure of Dau c 1 has been determined to a resolution of 2.7 A, revealing tight dimers. The structure of Dau c 1 is similar to those of the major allergens from celery, Api g 1, and birch pollen, Bet v 1. Electron density has been observed in the hydrophobic cavity of each monomer and has been modelled with polyethylene glycol oligomers of varying length. Comparison of the surface topology and physicochemical properties of Dau c 1 and Bet v 1 revealed that they may have some, but not all, epitopes in common. This is in agreement with the observation that the majority of carrot-allergic patients have Bet v 1 cross-reactive IgE antibodies, whereas others have Dau c 1-specific IgE antibodies which do not recognize Bet v 1.
Organizational Affiliation:
Core Program of Structural Biology and Biophysics, Biozentrum, University of Basel, CH-4056 Basel, Switzerland. zora.housley@unibas.ch