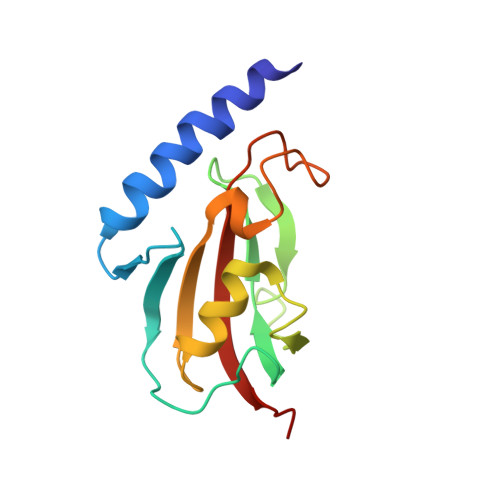
Solution structure of the Legionella pneumophila Mip-rapamycin complex.
Ceymann, A., Horstmann, M., Ehses, P., Schweimer, K., Paschke, A.K., Steinert, M., Faber, C.(2008) BMC Struct Biol 8: 17-17
- PubMed: 18366641
- DOI: https://doi.org/10.1186/1472-6807-8-17
- Primary Citation of Related Structures:
2VCD - PubMed Abstract:
Legionella pneumphila is the causative agent of Legionnaires' disease. A major virulence factor of the pathogen is the homodimeric surface protein Mip. It shows peptidyl-prolyl cis/trans isomerase activty and is a receptor of FK506 and rapamycin, which both inhibit its enzymatic function. Insight into the binding process may be used for the design of novel Mip inhibitors as potential drugs against Legionnaires' disease.
Organizational Affiliation:
Department of Experimental Physics 5, University of Würzburg, Würzburg, Germany. asceyman@physik.uni-wuerzburg.de