Structures of Thymidine Kinase 1 of Human and Mycoplasmic Origin
Welin, M., Kosinska, U., Mikkelsen, N.E., Carnrot, C., Zhu, C., Wang, L., Eriksson, S., Munch-Petersen, B., Eklund, H.(2004) Proc Natl Acad Sci U S A 101: 17970
- PubMed: 15611477
- DOI: https://doi.org/10.1073/pnas.0406332102
- Primary Citation of Related Structures:
1XBT, 2UZ3 - PubMed Abstract:
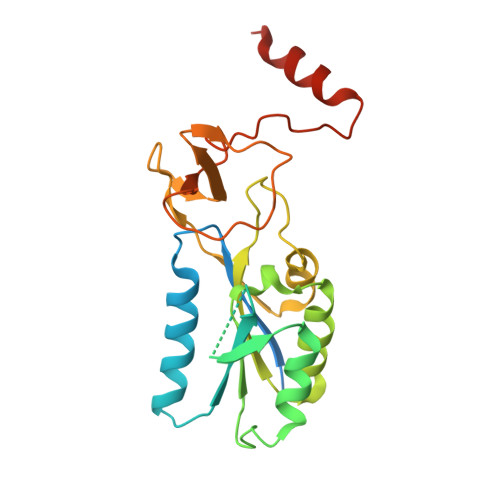
Cytosolic thymidine kinase 1, TK1, is a well known cell-cycle-regulated enzyme of importance in nucleotide metabolism as well as an activator of antiviral and anticancer drugs such as 3'-azido-3'-deoxythymidine (AZT). We have now determined the structures of the TK1 family, the human and Ureaplasma urealyticum enzymes, in complex with the feedback inhibitor dTTP. The TK1s have a tetrameric structure in which each subunit contains an alpha/beta-domain that is similar to ATPase domains of members of the RecA structural family and a domain containing a structural zinc. The zinc ion connects beta-structures at the root of a beta-ribbon that forms a stem that widens to a lasso-type loop. The thymidine of dTTP is hydrogen-bonded to main-chain atoms predominantly coming from the lasso loop. This binding is in contrast to other deoxyribonucleoside kinases where specific interactions occur with side chains. The TK1 structure differs fundamentally from the structures of the other deoxyribonucleoside kinases, indicating a different evolutionary origin.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, S-751 24 Uppsala, Sweden.