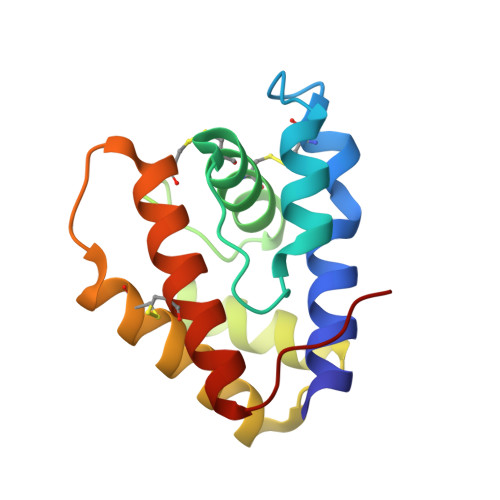
Bombyx mori pheromone-binding protein binding nonpheromone ligands: implications for pheromone recognition.
Lautenschlager, C., Leal, W.S., Clardy, J.(2007) Structure 15: 1148-1154
- PubMed: 17850754
- DOI: https://doi.org/10.1016/j.str.2007.07.013
- Primary Citation of Related Structures:
2P70, 2P71 - PubMed Abstract:
Insect pheromone-binding proteins (PBPs) transport sex pheromones through the aqueous layer surrounding G protein-coupled receptors that initiate signaling events leading to mating. This PBP-receptor system strongly discriminates between ligands with subtle structural differences, but it has proved difficult to distinguish the degree of discrimination of the PBP from that of the G protein-coupled receptor. The three-dimensional structures of the PBP of Bombyx mori, the silkworm moth, both with and without its cognate ligand bombykol ([E,Z]-10,12-hexadecadienol), have been determined by X-ray crystallography and NMR. In this paper, the structures of the same binding protein with bound iodohexadecane and bell pepper odorant were determined at 1.9 and 2.0 A, respectively. These structures illustrate the remarkable plasticity in the ligand binding site of the PBP, but suggest the protein might still act as a filter during pheromone signal processing.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA.