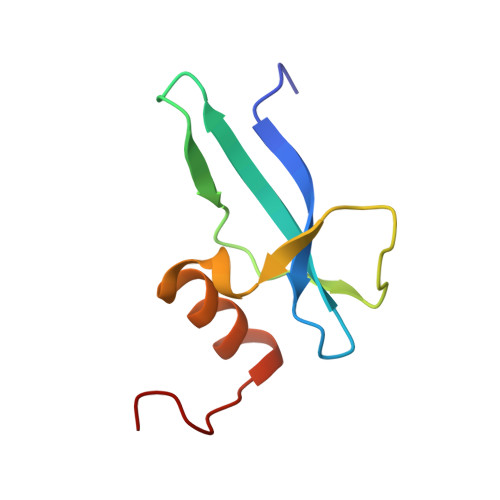
Structure determination of archaea-specific ribosomal protein L46a reveals a novel protein fold.
Feng, Y., Song, X., Lin, J., Xuan, J., Cui, Q., Wang, J.(2014) Biochem Biophys Res Commun 450: 67-72
- PubMed: 24875358
- DOI: https://doi.org/10.1016/j.bbrc.2014.05.077
- Primary Citation of Related Structures:
2MMP - PubMed Abstract:
Three archaea-specific ribosomal proteins recently identified show no sequence homology with other known proteins. Here we determined the structure of L46a, the most conserved one among the three proteins, from Sulfolobus solfataricus P2 using NMR spectroscopy. The structure presents a twisted β-sheet formed by the N-terminal part and two helices at the C-terminus. The L46a structure has a positively charged surface which is conserved in the L46a protein family and is the potential rRNA-binding site. Searching homologous structures in Protein Data Bank revealed that the structure of L46a represents a novel protein fold. The backbone dynamics identified by NMR relaxation experiments reveal significant flexibility at the rRNA binding surface. The potential position of L46a on the ribosome was proposed by fitting the structure into a previous electron microscopy map of the ribosomal 50S subunit, which indicated that L46a contacts to domain I of 23S rRNA near a multifunctional ribosomal protein L7ae.
Organizational Affiliation:
Shandong Provincial Key Laboratory of Energy Genetics, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, Shandong 266101, China. Electronic address: fengyg@qibebt.ac.cn.