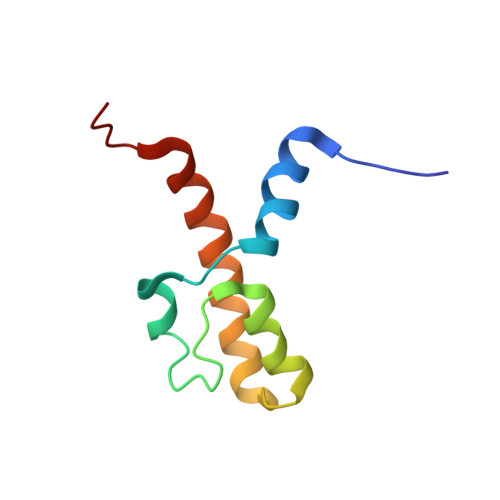
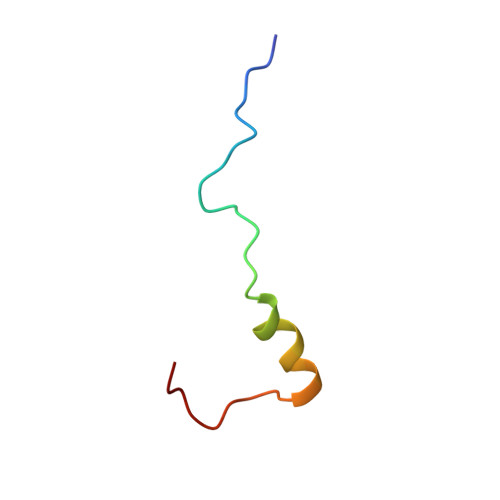
The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex.
Brzovic, P.S., Heikaus, C.C., Kisselev, L., Vernon, R., Herbig, E., Pacheco, D., Warfield, L., Littlefield, P., Baker, D., Klevit, R.E., Hahn, S.(2011) Mol Cell 44: 942-953
- PubMed: 22195967
- DOI: https://doi.org/10.1016/j.molcel.2011.11.008
- Primary Citation of Related Structures:
2LPB - PubMed Abstract:
The structural basis for binding of the acidic transcription activator Gcn4 and one activator-binding domain of the Mediator subunit Gal11/Med15 was examined by NMR. Gal11 activator-binding domain 1 has a four-helix fold with a small shallow hydrophobic cleft at its center. In the bound complex, eight residues of Gcn4 adopt a helical conformation, allowing three Gcn4 aromatic/aliphatic residues to insert into the Gal11 cleft. The protein-protein interface is dynamic and surprisingly simple, involving only hydrophobic interactions. This allows Gcn4 to bind Gal11 in multiple conformations and orientations, an example of a "fuzzy" complex, where the Gcn4-Gal11 interface cannot be described by a single conformation. Gcn4 uses a similar mechanism to bind two other unrelated activator-binding domains. Functional studies in yeast show the importance of residues at the protein interface, define the minimal requirements for a functional activator, and suggest a mechanism by which activators bind to multiple unrelated targets.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle, WA 98195, USA.