Solution structure of the dimerization domain of the eukaryotic stalk P1/P2 complex reveals the structural organization of eukaryotic stalk complex
Lee, K.-M., Yu, C.W.-H., Chiu, T.Y.-H., Sze, K.-H., Shaw, P.-C., Wong, K.-B.(2011) Nucleic Acids Res
- PubMed: 22135285
- DOI: https://doi.org/10.1093/nar/gkr1143
- Primary Citation of Related Structures:
2LBF - PubMed Abstract:
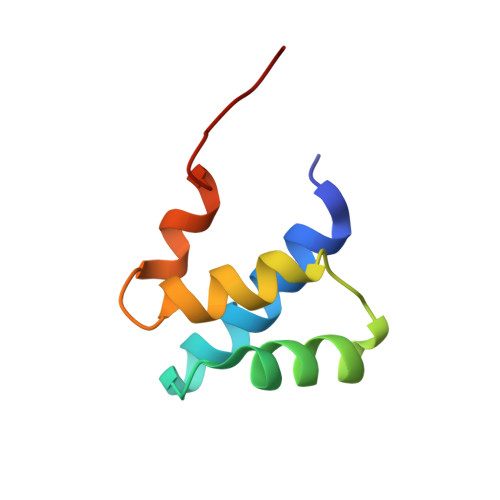
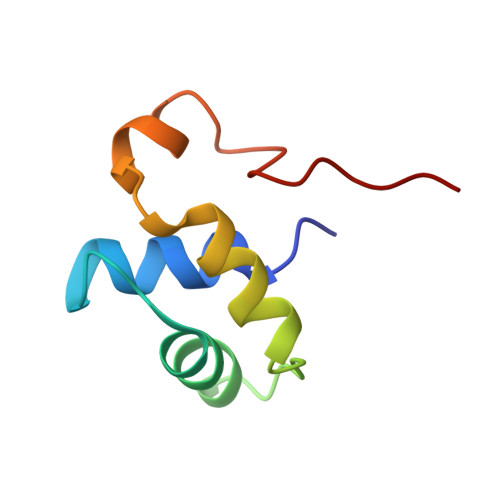
The lateral ribosomal stalk is responsible for the kingdom-specific binding of translation factors and activation of GTP hydrolysis during protein synthesis. The eukaryotic stalk is composed of three acidic ribosomal proteins P0, P1 and P2. P0 binds two copies of P1/P2 hetero-dimers to form a pentameric P-complex. The structure of the eukaryotic stalk is currently not known. To provide a better understanding on the structural organization of eukaryotic stalk, we have determined the solution structure of the N-terminal dimerization domain (NTD) of P1/P2 hetero-dimer. Helix-1, -2 and -4 from each of the NTD-P1 and NTD-P2 form the dimeric interface that buries 2200 A(2) of solvent accessible surface area. In contrast to the symmetric P2 homo-dimer, P1/P2 hetero-dimer is asymmetric. Three conserved hydrophobic residues on the surface of NTD-P1 are replaced by charged residues in NTD-P2. Moreover, NTD-P1 has an extra turn in helix-1, which forms extensive intermolecular interactions with helix-1 and -4 of NTD-P2. Truncation of this extra turn of P1 abolished the formation of P1/P2 hetero-dimer. Systematic truncation studies suggest that P0 contains two spine-helices that each binds one copy of P1/P2 hetero-dimer. Modeling studies suggest that a large hydrophobic cavity, which can accommodate the loop between the spine-helices of P0, can be found on NTD-P1 but not on NTD-P2 when the helix-4 adopts an 'open' conformation. Based on the asymmetric properties of NTD-P1/NTD-P2, a structural model of the eukaryotic P-complex with P2/P1:P1/P2 topology is proposed.
Organizational Affiliation:
School of Life Sciences, Centre for Protein Science and Crystallography, The Chinese University of Hong Kong, Hong Kong, China.