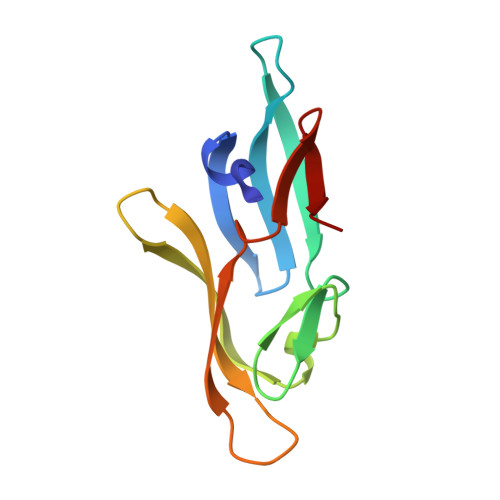
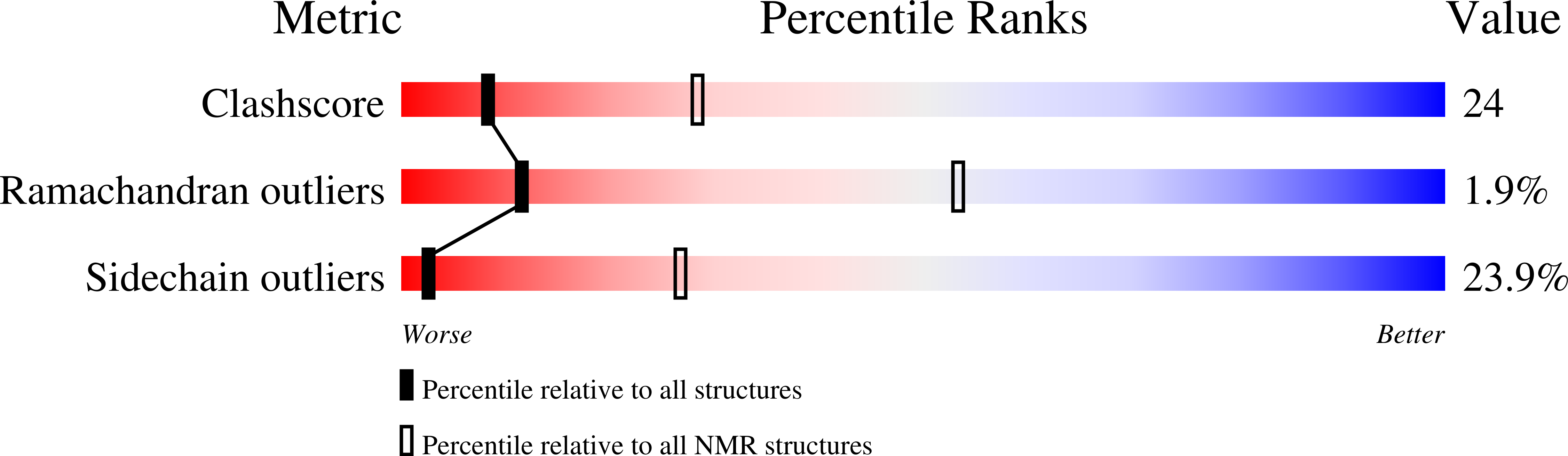NMR solution structure of a cyanovirin homolog from wheat head blight fungus.
Matei, E., Louis, J.M., Jee, J., Gronenborn, A.M.(2011) Proteins 79: 1538-1549
- PubMed: 21365681
- DOI: https://doi.org/10.1002/prot.22981
- Primary Citation of Related Structures:
2L2F - PubMed Abstract:
Members of the cyanovirin-N homolog (CVNH) lectin family are found in bacteria, fungi and plants. As part of our ongoing work on CVNH structure-function studies, we determined the high-resolution NMR solution structure of the homolog from the wheat head blight disease causing ascomycetous fungus Gibberella zeae (or Fusarium graminearum), hereafter called GzCVNH. Like cyanovirin-N (CV-N), GzCVNH comprises two tandem sequence repeats and the protein sequence exhibits 30% identity with CV-N. The overall structure is similar to those of other members of the CVNH family, with the conserved pseudo-symmetric halves of the structure, domains A and B, closely resembling recently determined structures of Tuber borchii, Neurospora crassa, and Ceratopteris richardii CVNH proteins. Although GzCVNH exhibits a similar glycan recognition profile to CV-N and specifically binds to Manα(1-2)Manα, its weak carbohydrate binding affinity to only one binding site is insufficient for conferring anti-HIV activity.
Organizational Affiliation:
Department of Structural Biology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania 15260, USA.