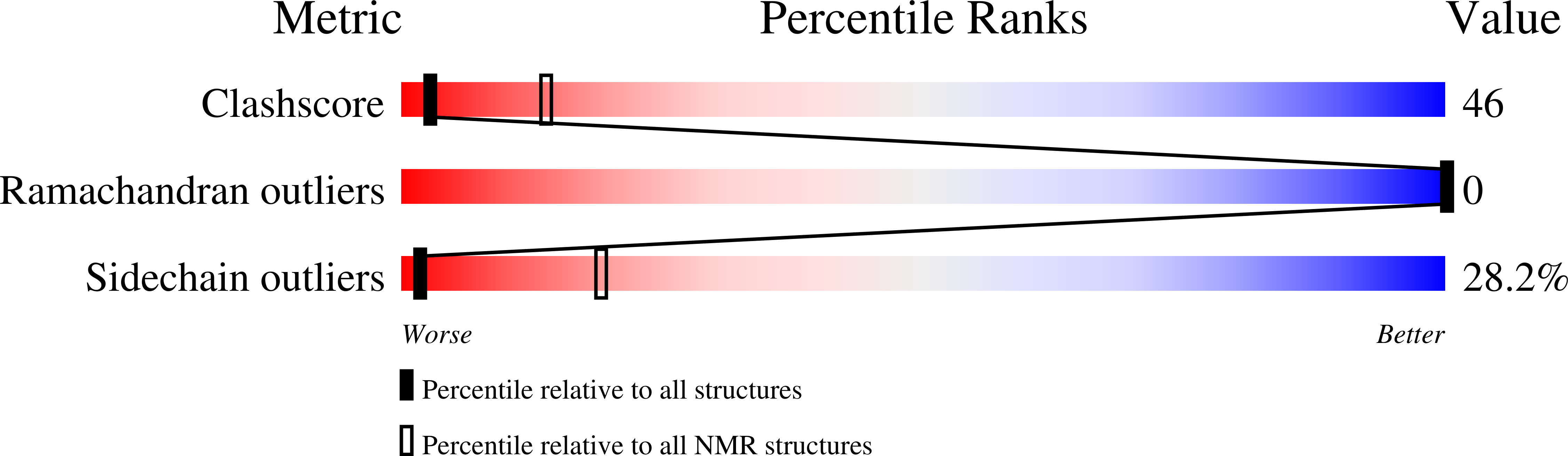Structure and mechanism of action of the antimicrobial peptide piscidin
Campagna, S., Saint, N., Molle, G., Aumelas, A.(2007) Biochemistry 46: 1771-1778
- PubMed: 17253775
- DOI: https://doi.org/10.1021/bi0620297
- Primary Citation of Related Structures:
2JOS - PubMed Abstract:
Piscidin, an antibacterial peptide isolated from the mast cells of striped bass, has potent antimicrobial activity against a broad spectrum of pathogens in vitro. We investigated the mechanism of action of this 22-residue cationic peptide by carrying out structural studies and electrophysiological experiments in lipid bilayers. Circular dichroism experiments showed that piscidin was unstructured in water but had a high alpha-helix content in dodecylphosphocholine (DPC) micelles. 1H NMR data in water and TFE confirmed these results and demonstrated that the segment of residues 8-17 adopted an alpha-helical structure in a micellar environment. This molecule has a marked amphipathic character, due to well-defined hydrophobic and hydrophilic sectors. This structure is similar to those determined for other cationic peptides involved in permeabilization of the bacterial membrane. Multichannel experiments with piscidin incorporated into azolectin planar bilayers gave reproducible I-V curves at various peptide concentrations and unambiguously showed that this peptide permeabilized the membrane. This pore forming activity was confirmed by single-channel experiments, with well-defined ion channels obtained at different voltages. The characteristics of the ion channels (voltage dependence, only one or two states of conductance) clearly suggest that piscidin is more likely to permeabilize the membrane by toroidal pore formation rather than via the "barrel-stave" mechanism.
Organizational Affiliation:
CNRS UMR5048, Centre de Biochimie Structurale, F34090 Montpellier, France.