Structure of the Tau60/Deltatau91 Subcomplex of Yeast Transcription Factor Iiic: Insights Into Preinitiation Complex Assembly
Mylona, A., Fernandez-Tornero, C., Legrand, P., Haupt, M., Sentenac, A., Acker, J., Muller, C.W.(2006) Mol Cell 24: 221
- PubMed: 17052456
- DOI: https://doi.org/10.1016/j.molcel.2006.08.013
- Primary Citation of Related Structures:
2J04 - PubMed Abstract:
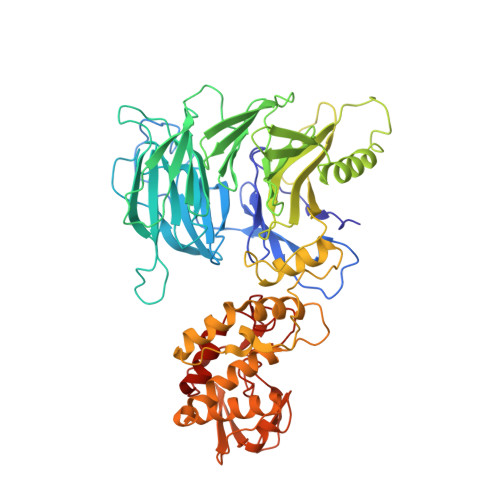
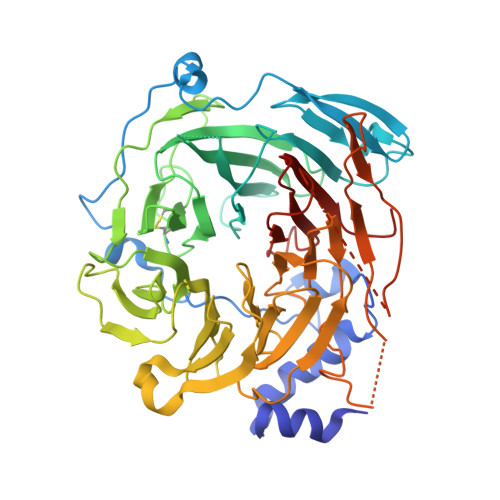
Yeast RNA polymerase III is recruited upon binding of subcomplexes tauA and tauB of transcription factor IIIC (TFIIIC) to the A and B blocks of tRNA gene promoters. The tauB subcomplex consists of subunits tau60, tau91, and tau138. We determined the 3.2 A crystal structure of tau60 bound to a large C-terminal fragment of tau91 (Deltatau91). Deltatau91 protein contains a seven-bladed propeller preceded by an N-terminal extension, whereas tau60 contains a structurally homologous propeller followed by a C-terminal domain with a novel alpha/beta fold. The two propeller domains do not have any detectable DNA binding activity and mediate heterodimer formation that may serve as scaffold for tau138 assembly. We show that the C-terminal tau60 domain interacts with the TATA binding protein (TBP). Recombinant tauB recruits TBP and stimulates TFIIIB-directed transcription on a TATA box containing tRNA gene, implying a combined contribution of tauA and tauB to preinitiation complex formation.
Organizational Affiliation:
European Molecular Biology Laboratory, Grenoble Outstation, B.P. 181, 38042 Grenoble Cedex 9, France.