Solution Structure of Human and Porcine Beta-Microseminoprotein
Ghasriani, H., Teilum, K., Johnsson, Y., Fernlund, P., Drakenberg, T.(2006) J Mol Biol 362: 502
- PubMed: 16930619
- DOI: https://doi.org/10.1016/j.jmb.2006.07.029
- Primary Citation of Related Structures:
2IZ3, 2IZ4 - PubMed Abstract:
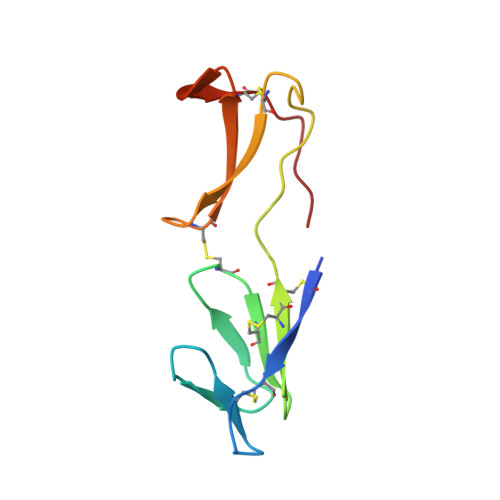
Beta-microseminoprotein (MSP) is a small cysteine-rich protein (molecular mass about 10 kDa) first isolated from human seminal plasma and later identified in several other organisms. The function of MSP is not known, but a recent study has shown MSP to bind CRISP-3, a protein present in neutrophilic granulocytes. The amino acid sequence is highly variable between species raising the question of the evolutionary conservation of the 3D structure. Here we present NMR solution structures of both the human and the porcine MSP. The two proteins (sequence identity 51%) have a very similar 3D structure with the secondary structure elements well conserved and with most of the amino acid substitutions causing a change of charge localized to one side of the molecule. MSP is a beta-sheet-rich protein with two distinct domains. The N-terminal domain is composed of a four-stranded beta-sheet, with the strands arranged according to the Greek key-motif, and a less structured part. The C-terminal domain contains two two-stranded beta-sheets with no resemblance to known structural motifs. The two domains, connected to each other by the peptide backbone, one disulfide bond, and interactions between the N and C termini, are oriented to give the molecule a rather extended structure. This global fold differs markedly from that of a previously published structure for porcine MSP, in which the two domains have an entirely different orientation to each other. The difference probably stems from a misinterpretation of ten specific inter-domain NOEs.
Organizational Affiliation:
Department of Biophysical Chemistry, Lund University, P.O. Box 124, SE-221 00 Lund, Sweden.