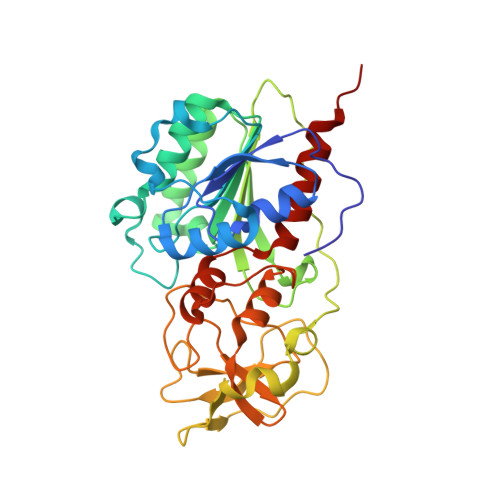
The Role of Arg165 Towards Base Flipping, Base Stabilization and Catalysis in M.HhaI.
Shieh, F.K., Youngblood, B., Reich, N.O.(2006) J Mol Biol 362: 516-527
- PubMed: 16926025
- DOI: https://doi.org/10.1016/j.jmb.2006.07.030
- Primary Citation of Related Structures:
2HR1, 2Z6Q - PubMed Abstract:
Arg165 forms part of a previously identified base flipping motif in the bacterial DNA cytosine methyltransferase, M.HhaI. Replacement of Arg165 with Ala has no detectable effect on either DNA or AdoMet affinity, yet causes the base flipping and restacking transitions to be decreased approximately 16 and 190-fold respectively, thus confirming the importance of this motif. However, these kinetic changes cannot account for the mutant's observed 10(5)-fold decreased catalytic rate. The mutant enzyme/cognate DNA cocrystal structure (2.79 A resolution) shows the target cytosine to be positioned approximately 30 degrees into the major groove, which is consistent with a major groove pathway for nucleotide flipping. The pyrimidine-sugar chi angle is rotated to approximately +171 degrees, from a range of -95 degrees to -120 degrees in B DNA, and -77 degrees in the WT M.HhaI complex. Thus, Arg165 is important for maintaining the cytosine positioned for nucleophilic attack by Cys81. The cytosine sugar pucker is in the C2'-endo-C3'-exo (South conformation), in contrast to the previously reported C3'-endo (North conformation) described for the original 2.70 A resolution cocrystal structure of the WT M.HhaI/DNA complex. We determined a high resolution structure of the WT M.HhaI/DNA complex (1.96 A) to better determine the sugar pucker. This new structure is similar to the original, lower resolution WT M.HhaI complex, but shows that the sugar pucker is O4'-endo (East conformation), intermediate between the South and North conformers. In summary, Arg165 plays significant roles in base flipping, cytosine positioning, and catalysis. Furthermore, the previously proposed M.HhaI-mediated changes in sugar pucker may not be an important contributor to the base flipping mechanism. These results provide insights into the base flipping and catalytic mechanisms for bacterial and eukaryotic DNA methyltransferases.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Santa Barbara, CA 93106-9510, USA.