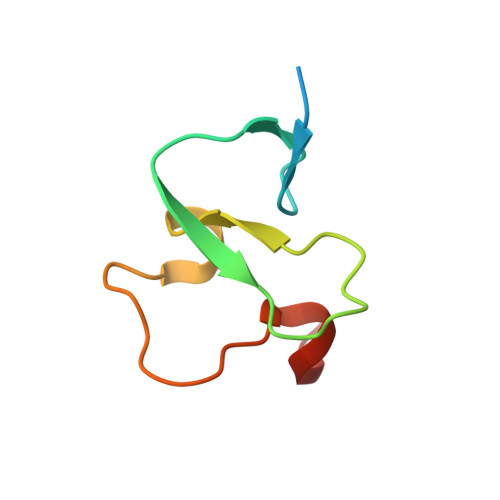
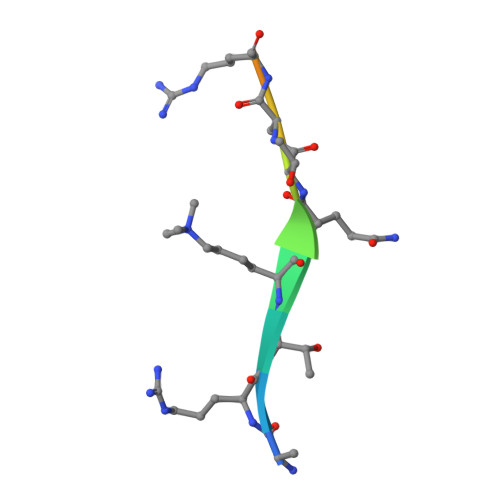
Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2.
Pena, P.V., Davrazou, F., Shi, X., Walter, K.L., Verkhusha, V.V., Gozani, O., Zhao, R., Kutateladze, T.G.(2006) Nature 442: 100-103
- PubMed: 16728977
- DOI: https://doi.org/10.1038/nature04814
- Primary Citation of Related Structures:
2G6Q - PubMed Abstract:
Covalent modifications of histone tails have a key role in regulating chromatin structure and controlling transcriptional activity. In eukaryotes, histone H3 trimethylated at lysine 4 (H3K4me3) is associated with active chromatin and gene expression. We recently found that plant homeodomain (PHD) finger of tumour suppressor ING2 (inhibitor of growth 2) binds H3K4me3 and represents a new family of modules that target this epigenetic mark. The molecular mechanism of H3K4me3 recognition, however, remains unknown. Here we report a 2.0 A resolution structure of the mouse ING2 PHD finger in complex with a histone H3 peptide trimethylated at lysine 4. The H3K4me3 tail is bound in an extended conformation in a deep and extensive binding site consisting of elements that are conserved among the ING family of proteins. The trimethylammonium group of Lys 4 is recognized by the aromatic side chains of Y215 and W238 residues, whereas the intermolecular hydrogen-bonding and complementary surface interactions, involving Ala 1, Arg 2, Thr 3 and Thr 6 of the peptide, account for the PHD finger's high specificity and affinity. Substitution of the binding site residues disrupts H3K4me3 interaction in vitro and impairs the ability of ING2 to induce apoptosis in vivo. Strong binding of other ING and YNG PHD fingers suggests that the recognition of H3K4me3 histone code is a general feature of the ING/YNG proteins. Elucidation of the mechanisms underlying this novel function of PHD fingers provides a basis for deciphering the role of the ING family of tumour suppressors in chromatin regulation and signalling.
Organizational Affiliation:
Department of Pharmacology, University of Colorado Health Sciences Center, Aurora, Colorado 80045, USA.