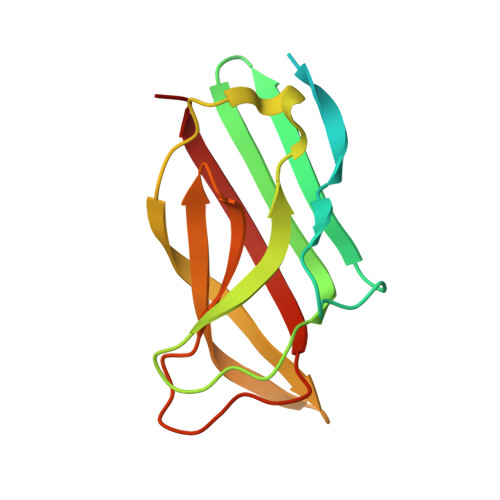
Crystal Structure of the Receptor-Binding Protein Head Domain from Lactococcus lactis Phage bIL170
Ricagno, S., Campanacci, V., Blangy, S., Spinelli, S., Tremblay, D., Moineau, S., Tegoni, M., Cambillau, C.(2006) J Virol 80: 9331-9335
- PubMed: 16940545
- DOI: https://doi.org/10.1128/JVI.01160-06
- Primary Citation of Related Structures:
2FSD - PubMed Abstract:
Lactococcus lactis, a gram-positive bacterium widely used by the dairy industry, is subject to lytic phage infections. In the first step of infection, phages recognize the host saccharidic receptor using their receptor binding protein (RBP). Here, we report the 2.30-A-resolution crystal structure of the RBP head domain from phage bIL170. The structure of the head monomer is remarkably close to those of other lactococcal phages, p2 and TP901-1, despite any sequence identity with them. The knowledge of the three-dimensional structures of three RBPs gives a better insight into the module exchanges which have occurred among phages.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques, UMR 6098 CNRS and Universités Aix-Marseille I & II, Campus de Luminy, Case 932, 13288 Marseille Cedex 09, France.