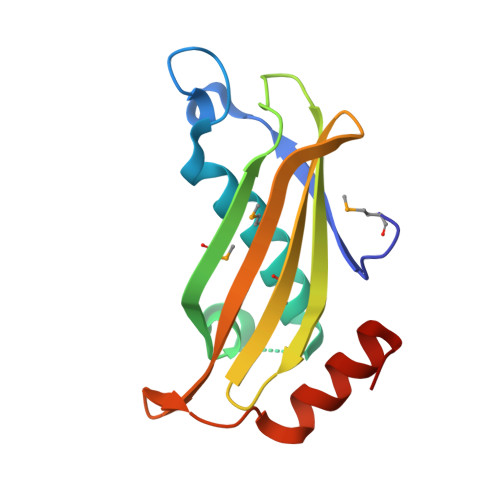
Structure of the putative thioesterase protein TTHA1846 from Thermus thermophilus HB8 complexed with coenzyme A and a zinc ion.
Hosaka, T., Murayama, K., Kato-Murayama, M., Urushibata, A., Akasaka, R., Terada, T., Shirouzu, M., Kuramitsu, S., Yokoyama, S.(2009) Acta Crystallogr D Biol Crystallogr 65: 767-776
- PubMed: 19622860
- DOI: https://doi.org/10.1107/S0907444909015601
- Primary Citation of Related Structures:
2CYE - PubMed Abstract:
TTHA1846 is a conserved hypothetical protein from Thermus thermophilus HB8 with a molecular mass of 15.1 kDa that belongs to the thioesterase superfamily (Pfam 03061). Here, the 1.9 A resolution crystal structure of TTHA1846 from T. thermophilus is reported. The crystal structure is a dimer of dimers. Each subunit adopts the so-called hot-dog fold composed of five antiparallel beta-strands flanked on one side by a rather long alpha-helix and shares structural similarity to a number of thioesterases. Unexpectedly, TTHA1846 binds one metal ion and one ligand per subunit. The ligand density was modelled as coenzyme A (CoA). Its structure was confirmed by MALDI-TOF mass spectrometry and electron-density mapping. X-ray absorption fine-structure (XAFS) measurement of the crystal unambiguously characterized the metal ion as zinc. The zinc ion is tetrahedrally coordinated by the side chains of Asp18, His22 and Glu50 and the CoA thiol group. This is the first structural report of the interaction of CoA with a zinc ion. From structural and database analyses, it was speculated that the zinc ion may play an inhibitory role in the enzymatic activity.
Organizational Affiliation:
Protein Research Group, RIKEN Systems and Structural Biology Center, Tsurumi-ku, Yokohama 230-0045, Japan.