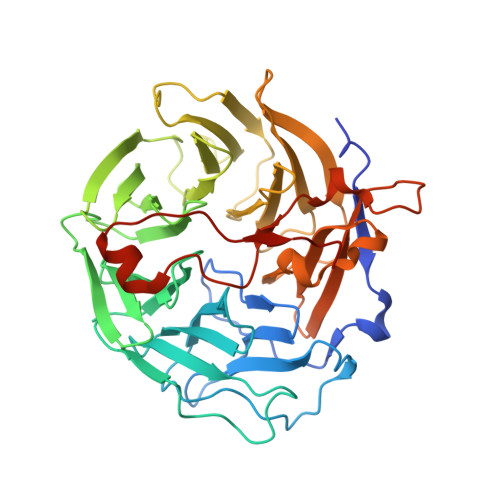
The crystal structure of murine coronin-1: a regulator of actin cytoskeletal dynamics in lymphocytes
Appleton, B.A., Wu, P., Wiesmann, C.(2006) Structure 14: 87-96
- PubMed: 16407068
- DOI: https://doi.org/10.1016/j.str.2005.09.013
- Primary Citation of Related Structures:
2AQ5, 2B4E - PubMed Abstract:
Mammalian coronin-1 is preferentially expressed in hematopoietic cells and plays a poorly understood role in the dynamic reorganization of the actin cytoskeleton. Sequence analysis of coronin-1 revealed five WD40 repeats that were predicted to form a beta propeller. They are followed by a 130 residue extension and a 30 residue leucine zipper domain that is responsible for multimerization of the protein. Here, we present the crystal structure of murine coronin-1 without the leucine zipper at 1.75 A resolution. Coronin-1 forms a seven-bladed beta propeller composed of the five predicted WD40 repeats and two additional blades that lack any homology to the canonical WD40 motif. The C-terminal extension adopts an extended conformation, packs tightly against the bottom surface of the propeller, and is likely to be required for the structural stability of the propeller. Analysis of charged and conserved surface residues delineate possible binding sites for F-actin on the beta propeller.
Organizational Affiliation:
Department of Protein Engineering, Genentech, Inc., South San Francisco, California 94080, USA.