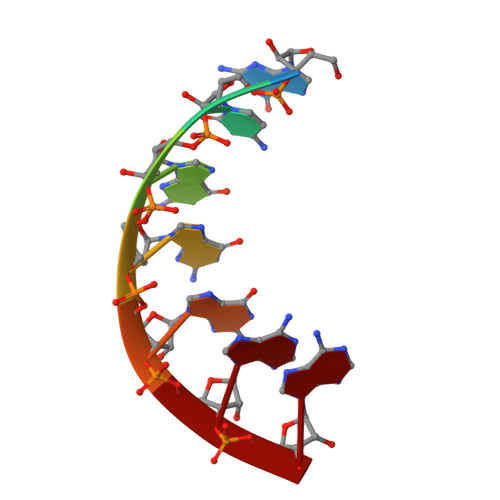
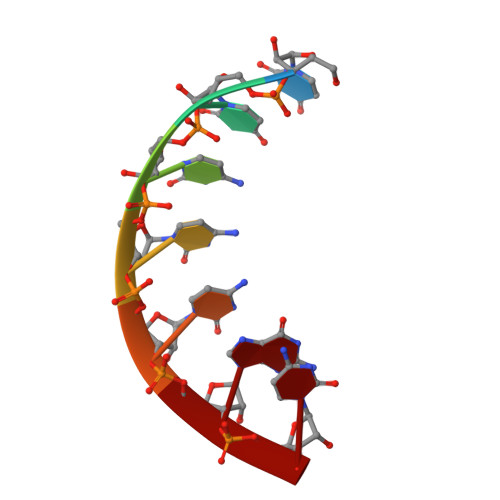
Crystal Structure of an Escherichia Coli Trnagly Microhelix at 2.0 Angstrom Resolution
Forster, C., Brauer, A.B.E., Perbandt, M., Lehmann, D., Furste, J.P., Betzel, C., Erdmann, V.A.(2007) Biochem Biophys Res Commun 363: 621
- PubMed: 17888869
- DOI: https://doi.org/10.1016/j.bbrc.2007.09.008
- Primary Citation of Related Structures:
2VAL - PubMed Abstract:
tRNA identity elements determine the correct aminoacylation by the cognate aminoacyl-tRNA synthetase. In class II aminoacyl tRNA synthetase systems, tRNA specificity is assured by rather few and simple recognition elements, mostly located in the acceptor stem of the tRNA. Here we present the crystal structure of an Escherichia coli tRNA(Gly) aminoacyl stem microhelix at 2.0 A resolution. The tRNA(Gly) microhelix crystallizes in the space group P3(2)21 with the cell constants a=b=35.35 A, c=130.82 A, gamma=120 degrees . The helical parameters, solvent molecules and a potential magnesium binding site are discussed.
Organizational Affiliation:
Institute of Chemistry and Biochemistry, Free University Berlin, Thielallee 63, 14195 Berlin, Germany.