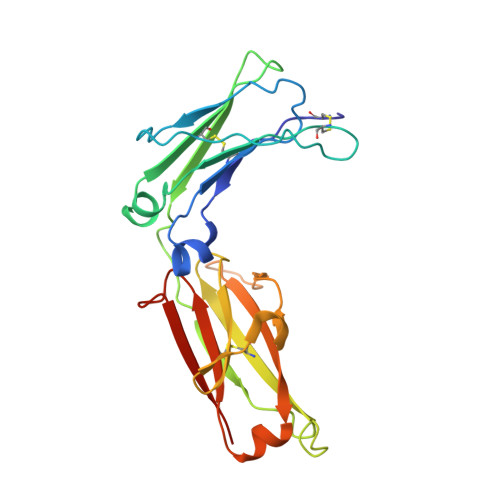
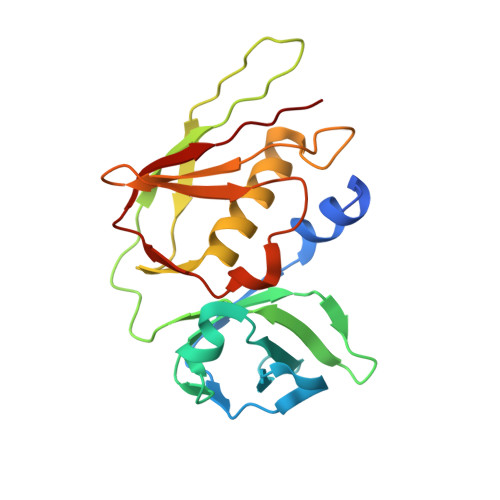
Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1
Ramsland, P.A., Willoughby, N., Trist, H.M., Farrugia, W., Hogarth, P.M., Fraser, J.D., Wines, B.D.(2007) Proc Natl Acad Sci U S A 104: 15051-15056
- PubMed: 17848512
- DOI: https://doi.org/10.1073/pnas.0706028104
- Primary Citation of Related Structures:
2QEJ - PubMed Abstract:
Infection by Staphylococcus aureus can result in severe conditions such as septicemia, toxic shock, pneumonia, and endocarditis with antibiotic resistance and persistent nasal carriage in normal individuals being key drivers of the medical impact of this virulent pathogen. In both virulent infection and nasal colonization, S. aureus encounters the host immune system and produces a wide array of factors that frustrate host immunity. One in particular, the prototypical staphylococcal superantigen-like protein SSL7, potently binds IgA and C5, thereby inhibiting immune responses dependent on these major immune mediators. We report here the three-dimensional structure of the complex of SSL7 with Fc of human IgA1 at 3.2 A resolution. Two SSL7 molecules interact with the Fc (one per heavy chain) primarily at the junction between the Calpha2 and Calpha3 domains. The binding site on each IgA chain is extensive, with SSL7 shielding most of the lateral surface of the Calpha3 domain. However, the SSL7 molecules are positioned such that they should allow binding to secretory IgA. The key IgA residues interacting with SSL7 are also bound by the leukocyte IgA receptor, FcalphaRI (CD89), thereby explaining how SSL7 potently inhibits IgA-dependent cellular effector functions mediated by FcalphaRI, such as phagocytosis, degranulation, and respiratory burst. Thus, the ability of S. aureus to subvert IgA-mediated immunity is likely to facilitate survival in mucosal environments such as the nasal passage and may contribute to systemic infections.
Organizational Affiliation:
The Inflammatory Disease and Structural Immunology Laboratories, The Burnet Institute, Austin Hospital, Studley Road, Heidelberg, Victoria 3084, Australia.