Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
Hoang, H.N., Song, K., Hill, T.A., Derksen, D.R., Edmonds, D.J., Kok, W.M., Limberakis, C., Liras, S., Loria, P.M., Mascitti, V., Mathiowetz, A.M., Mitchell, J.M., Piotrowski, D.W., Price, D.A., Stanton, R.V., Suen, J.Y., Withka, J.M., Griffith, D.A., Fairlie, D.P.(2015) J Med Chem 58: 4080-4085
- PubMed: 25839426
- DOI: https://doi.org/10.1021/acs.jmedchem.5b00166
- Primary Citation of Related Structures:
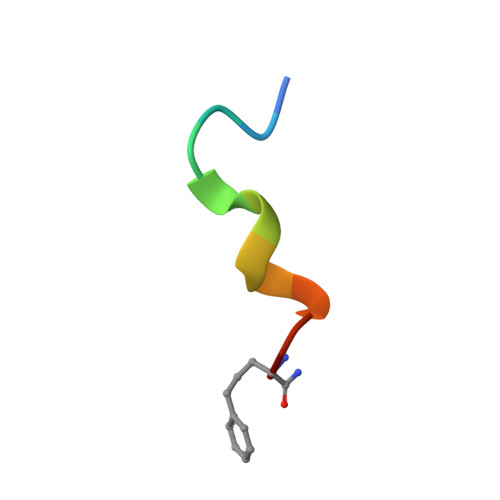
2N08, 2N09, 2N0I, 2N0N - PubMed Abstract:
Cyclic constraints are incorporated into an 11-residue analogue of the N-terminus of glucagon-like peptide-1 (GLP-1) to investigate effects of structure on agonist activity. Cyclization through linking side chains of residues 2 and 5 or 5 and 9 produced agonists at nM concentrations in a cAMP assay. 2D NMR and CD spectra revealed an N-terminal β-turn and a C-terminal helix that differentially influenced affinity and agonist potency. These structures can inform development of small molecule agonists of the GLP-1 receptor to treat type 2 diabetes.
Organizational Affiliation:
†Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland 4072, Australia.