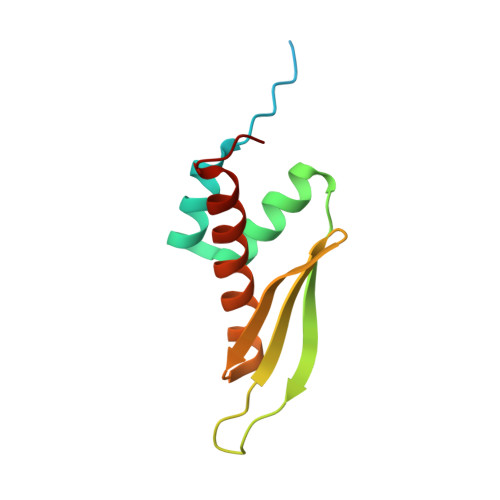
A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1.
Barraud, P., Banerjee, S., Mohamed, W.I., Jantsch, M.F., Allain, F.H.(2014) Proc Natl Acad Sci U S A 111: E1852-E1861
- PubMed: 24753571
- DOI: https://doi.org/10.1073/pnas.1323698111
- Primary Citation of Related Structures:
2MDR - PubMed Abstract:
The human RNA-editing enzyme adenosine deaminase acting on RNA (ADAR1) carries a unique nuclear localization signal (NLS) that overlaps one of its double-stranded RNA-binding domains (dsRBDs). This dsRBD-NLS is recognized by the nuclear import receptor transportin 1 (Trn1; also called karyopherin-β2) in an RNA-sensitive manner. Most Trn1 cargos bear a well-characterized proline-tyrosine-NLS, which is missing from the dsRBD-NLS. Here, we report the structure of the dsRBD-NLS, which reveals an unusual dsRBD fold extended by an additional N-terminal α-helix that brings the N- and C-terminal flanking regions in close proximity. We demonstrate experimentally that the atypical ADAR1-NLS is bimodular and is formed by the combination of the two flexible fragments flanking the folded domain. The intervening dsRBD acts only as an RNA-sensing scaffold, allowing the two NLS modules to be properly positioned for interacting with Trn1. We also provide a structural model showing how Trn1 can recognize the dsRBD-NLS and how dsRNA binding can interfere with Trn1 binding.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, Eidgenössiche Technische Hochschule Zürich, CH-8093 Zürich, Switzerland.